Search Count: 18
 |
Mechanistic Insights Into The Versatile Stoichiometry And Biased Signaling Of The Apelin Receptor-Arrestin Complex
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: A1L6R, CLR |
 |
Cryo-Em Structure Of Dimeric Apjr And Two Beta-Arrestins Complex With Small Molecules
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: A1L6R, CLR |
 |
Cryo-Em Structure Of Dimeric Apjr And One Beta-Arrestin Complex With Small Molecules
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: A1L6R |
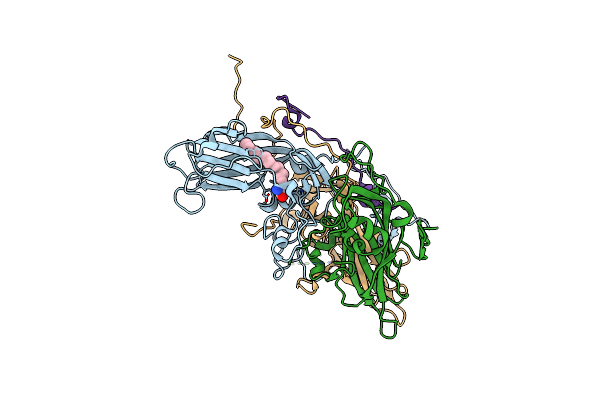 |
Cryo-Em Structure Of Enterovirus A71 Mature Virion In Complex With Fab H1A6.2
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS Ligands: SPH |
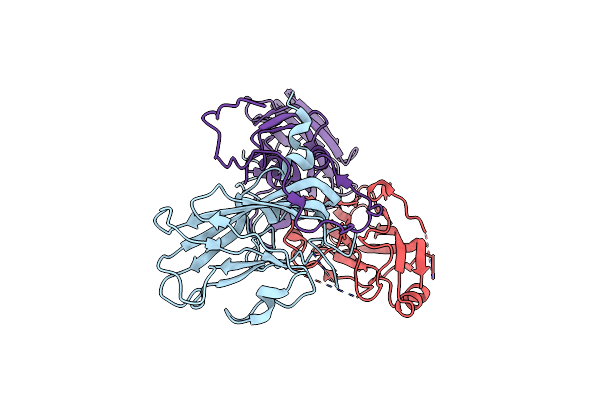 |
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
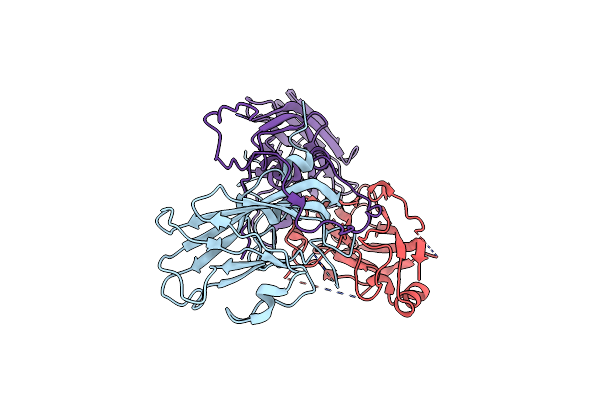 |
Cryo-Em Structure Of Enterovirus A71 Empty Particle In Complex With Fab H1A6.2
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
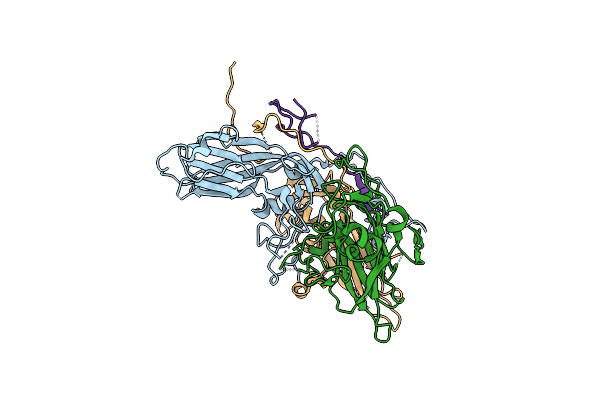 |
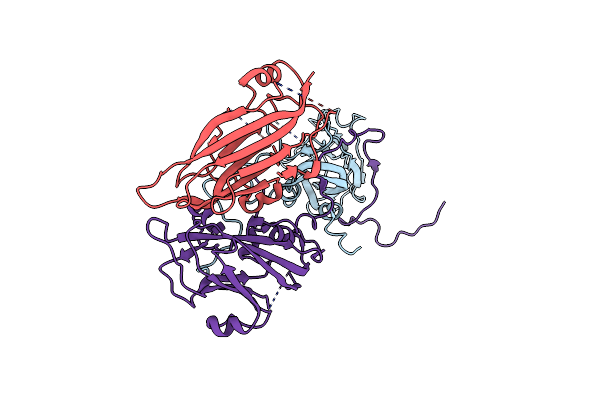
Cryo-Em Structure Of Coxsackievirus A16 Mature Virion In Complex With Fab H1A6.2
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
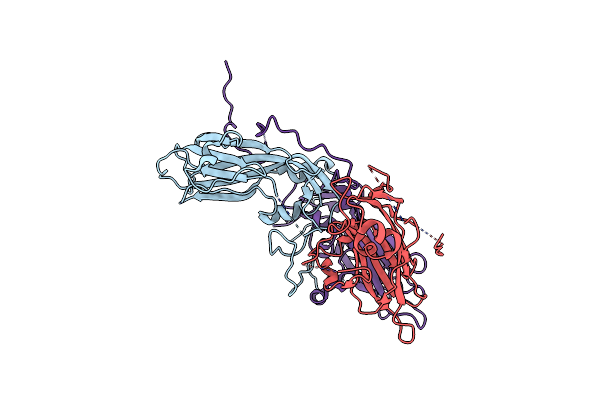 |
Cryo-Em Structure Of Coxsackievirus A16 A-Particle In Complex With Fab H1A6.2
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
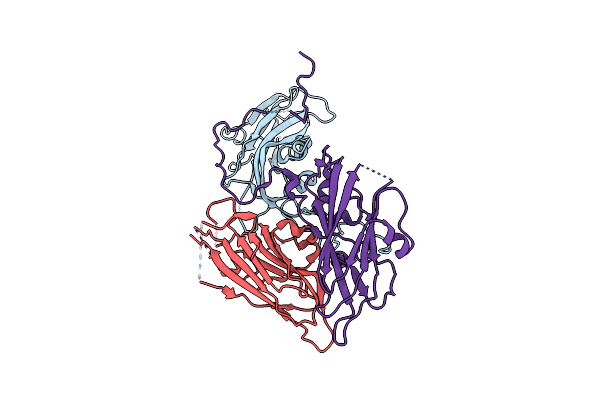 |
Cryo-Em Structure Of Coxsackievirus A16 Empty Particle In Complex With Fab H1A6.2
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
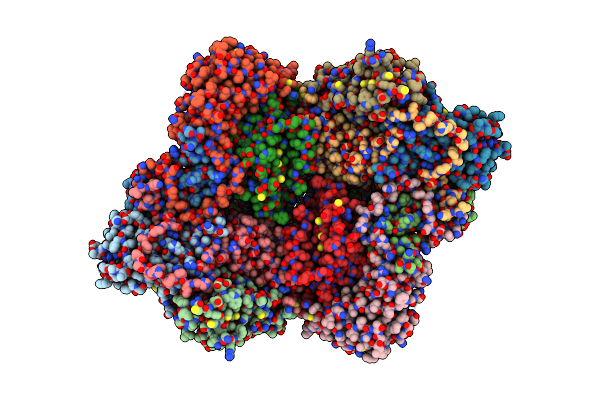 |
Cryo-Em Structure Of Coxsackievirus A16 Empty Particle In Complex With Fab H1A6.2 (Local Refinement)
Organism: Mus musculus, Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRAL PROTEIN |
 |
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
 |
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS Ligands: SPH |
 |
Organism: Scylla serrata reovirus sz-2007
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: VIRUS |
 |
Organism: Scylla serrata reovirus sz-2007
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: VIRUS |
 |
 |
Cryo-Em Structure Of Mud Crab Tombus-Like Virus At 3.3 Angstroms Resolution
Organism: Wenzhou tombus-like virus 18
Method: ELECTRON MICROSCOPY Release Date: 2019-01-16 Classification: VIRUS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.23 Å Release Date: 2018-12-19 Classification: transferase/transferase inhibitor Ligands: ZN, KHM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2017-09-27 Classification: transferase/transferase inhibitor Ligands: ZN, 8ZM |

