Search Count: 29
 |
Structure Of The Acyltransferase Domain Of Ncde, A Multi-Domain Nrps Protein From Nocardichelin Biosynthesis
Organism: Nocardia carnea
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: EPE, GOL |
 |
Structure Of E1277A Mutant Of The Acyltransferase Domain Of Ncde, A Multi-Domain Nrps Protein From Nocardichelin Biosynthesis
Organism: Nocardia carnea
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: EPE, GOL |
 |
Structure Of A Y1216F Mutant Of The Acyltransferase Domain Of Ncde, A Multi-Domain Nrps Protein From Nocardichelin Biosynthesis
Organism: Nocardia carnea
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: EPE |
 |
Structure Of A K1305A Mutant Of The Acyltransferase Domain Of Ncde, A Multi-Domain Nrps Protein From Nocardichelin Biosynthesis
Organism: Nocardia carnea
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: EPE |
 |
Organism: Nocardia carnea nbrc 14403
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: MLI |
 |
Crystal Structure Of Tet(X7) Bound To Anhydrotetracycline C10-Benzoate Ester
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-03-05 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: A1BDP, FAD |
 |
Structure Of E. Coli Dihydrofolate Reductase (Dhfr) In An Occluded Conformation And In Complex With A Cycloguanil Derivative
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: A1A3E |
 |
Structure Of E. Coli Dihydrofolate Reductase (Dhfr) In An Occluded Conformation And In Complex With Cycloguanil
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: 1CY, FLC |
 |
Structure Of Fbsh, An Nrps Adenylation Domain In The Fimsbactin Biosynthetic Pathway Bound To 2,3-Dihydroxybenzoic Acid.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-20 Classification: LIGASE/SUBSTRATE Ligands: DBH, EDO |
 |
Structure Of Fbsh, An Nrps Adenylation Domain In The Fimsbactin Biosynthetic Pathway Bound To Salicyl-Ams
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-11-20 Classification: LIGASE/INHIBITOR Ligands: KT0, EDO |
 |
Organism: Legionella massiliensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Legionella clemsonensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
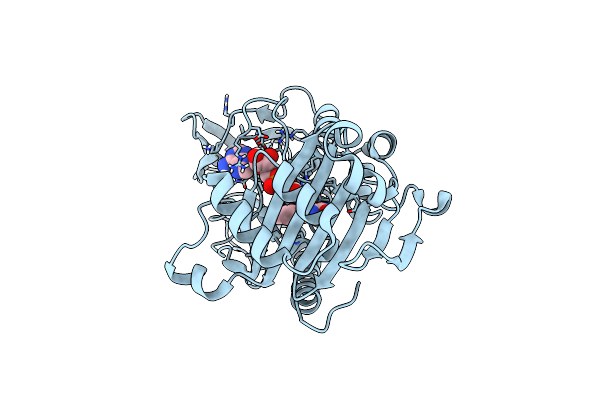
Organism: Chryseobacterium oncorhynchi
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: TDC, FAD |
 |
Organism: Chryseobacterium oncorhynchi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE Ligands: FAD |
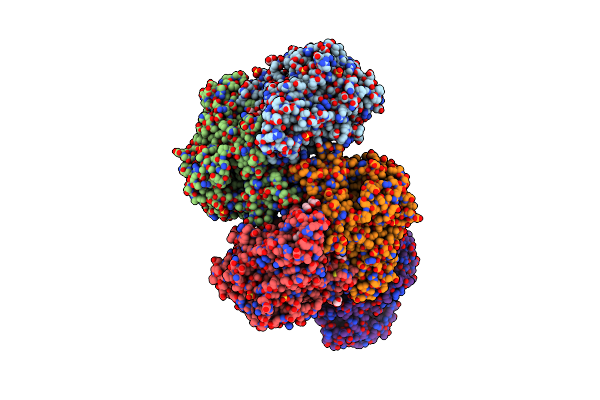 |
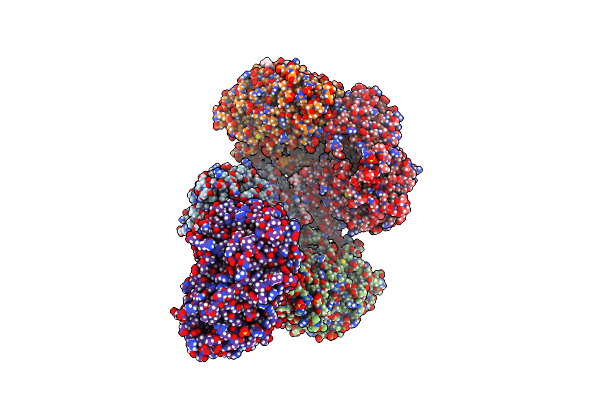
Crystal Structure Of Desd, The Desferrioxamine Synthetase From The Streptomyces Griseoflavus Ferrimycin Biosynthetic Pathway
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-07-06 Classification: LIGASE Ligands: GOL, SO4 |
 |
Crystal Structure Of Atp Bound Desd, The Desferrioxamine Synthetase From The Streptomyces Griseoflavus Ferrimycin Biosynthetic Pathway
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-07-06 Classification: LIGASE Ligands: SO4, ATP, MG, B3P, MPD |
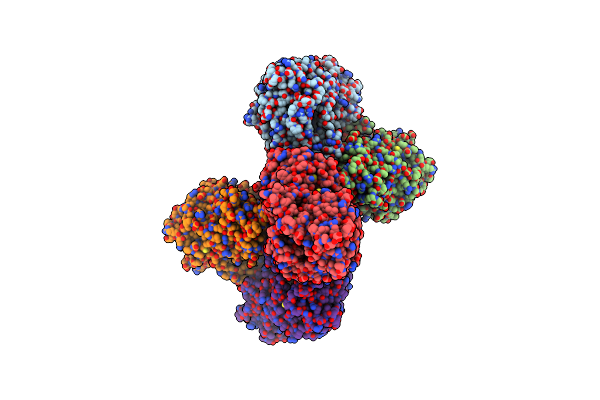 |
Crystal Structure Of Amp+Ppi Bound Desd, The Desferrioxamine Synthetase From The Streptomyces Griseoflavus Ferrimycin Biosynthetic Pathway
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2022-07-06 Classification: LIGASE Ligands: GOL, PPV, AMP, SO4, MG |
 |
Crystal Structure Of Hsc-Ams Bound Desd, The Desferrioxamine Synthetase From The Streptomyces Griseoflavus Ferrimycin Biosynthetic Pathway
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-07-06 Classification: LIGASE/INHIBITOR Ligands: GOL, I3U, LMS, PO4, MG, SO4 |
 |
Crystal Structure Of Desd, The Desferrioxamine Synthetase From The Streptomyces Violaceus Salmycin Biosynthetic Pathway
Organism: Streptomyces violaceus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-07-06 Classification: LIGASE Ligands: EDO, MG, FLC |
 |
Structure Of The Siderophore Interacting Protein From Acinetbacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: FAD |

