Search Count: 108
 |
A Double Mutant Of Serratia Marcescens Hemophore Receptor Hasr In Complex With Its Hemophore Hasa And Heme
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-06-29 Classification: TRANSPORT PROTEIN Ligands: HEM |
 |
Structure Of Trmbl2, An Archaeal Chromatin Protein, Shows A Novel Mode Of Dna Binding.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-09-02 Classification: DNA BINDING PROTEIN Ligands: GOL, MPD |
 |
Structure Of Trmbl2, An Archaeal Chromatin Protein, Shows A Novel Mode Of Dna Binding.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-09-02 Classification: DNA BINDING PROTEIN Ligands: GOL, IMD |
 |
Structure Of Trmbl2, An Archaeal Chromatin Protein, Shows A Novel Mode Of Dna Binding.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-09-02 Classification: DNA BINDING PROTEIN Ligands: GOL |
 |
Structure Of Trmbl2, An Archaeal Chromatin Protein, Shows A Novel Mode Of Dna Binding.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-09-02 Classification: DNA BINDING PROTEIN Ligands: CA |
 |
Chorismatase Mechanisms Reveal Fundamentally Different Types Of Reaction In A Single Conserved Protein Fold
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-08-19 Classification: OXIDOREDUCTASE Ligands: 3HB, SO4 |
 |
Chorismatase Mechanisms Reveal Fundamentally Different Types Of Reaction In A Single Conserved Protein Fold
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-08-19 Classification: OXIDOREDUCTASE Ligands: 3EB, SO4, PEG |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-02-11 Classification: GENE REGULATION |
 |
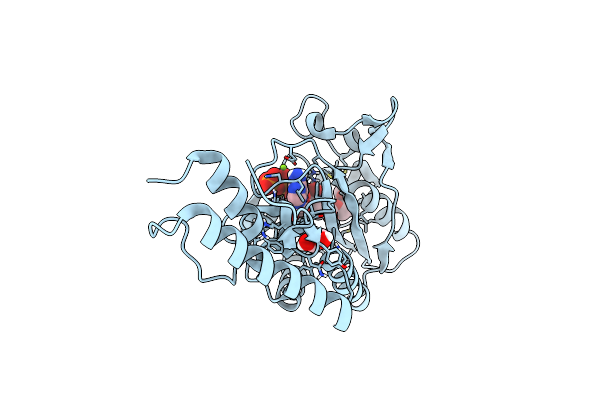
Catalytic Domain Of Pyrrolysyl-Trna Synthetase Mutant Y306A, Y384F In Its Apo Form
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-04-30 Classification: LIGASE Ligands: PEG, EDO |
 |
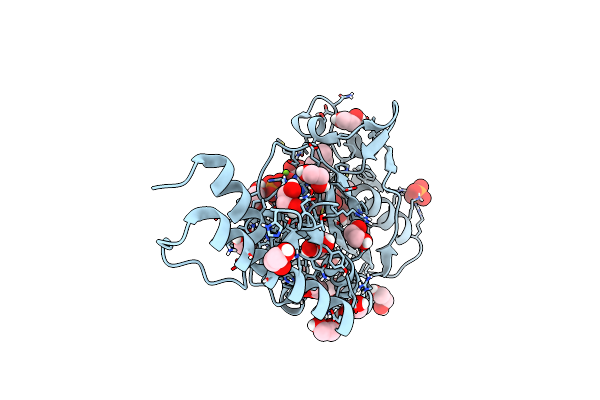
Catalytic Domain Of Pyrrolysyl-Trna Synthetase Mutant Y306A, Y384F In Complex With An Adenylated Furan-Bearing Noncanonical Amino Acid And Pyrophosphate
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-04-30 Classification: LIGASE Ligands: LYS, AMP, FU0, POP, MG, EDO |
 |
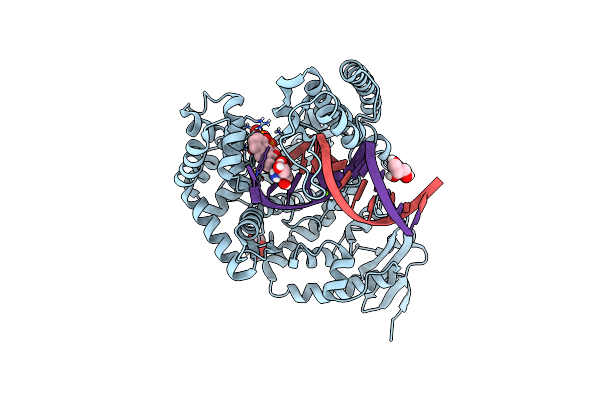
Catalytic Domain Of Pyrrolysyl-Trna Synthetase Mutant Y306A, Y384F In Complex With Amppnp
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-04-30 Classification: LIGASE Ligands: ANP, EDO, MG, AXZ, PO4 |
 |
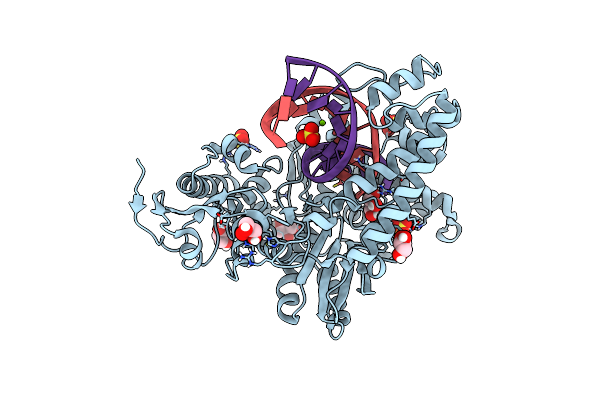
Crystal Structure Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus In A Partially Closed Complex With The Artificial Base Pair D5Sics-Dnamtp
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2013-12-11 Classification: TRANSFERASE/DNA Ligands: GOL, BMR, MG, TRS |
 |
Binary Complex Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus With The Artificial Base Pair Dnam-D5Sics At The Postinsertion Site (Sequence Context 1)
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-12-11 Classification: TRANSFERASE/DNA Ligands: GOL, SO4, MG |
 |
Binary Complex Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus With The Aritificial Base Pair D5Sics-Dnam At The Postinsertion Site (Sequence Context 2)
Organism: Synthetic construct, Thermus aquaticus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2013-12-11 Classification: TRANSFERASE/DNA Ligands: SO4, GOL, MG |
 |
Binary Complex Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus With The Aritificial Base Pair Dnam-D5Sics At The Postinsertion Site (Sequence Context 3)
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2013-12-11 Classification: TRANSFERASE/DNA Ligands: SO4, PGE |
 |
Binary Complex Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus With The Aritificial Base Pair Dnam-D5Sics At The Postinsertion Site (Sequence Context 2)
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-12-11 Classification: TRANSFERASE/DNA Ligands: SO4, MG, CL, TRS |
 |
Crystal Structure Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus In An Open Binary Complex With D5Sics As Templating Nucleotide
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2013-12-11 Classification: TRANSFERASE/DNA Ligands: FMT, MG, GOL |
 |
Organism: Streptomyces hygroscopicus subsp. ascomyceticus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2013-09-18 Classification: HYDROLASE Ligands: 3EB |
 |
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-09-18 Classification: TRANSFERASE/DNA Ligands: DCT, MG |
 |
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-09-18 Classification: TRANSFERASE/DNA/RNA Ligands: DCT, CL, MG, FMT |

