Search Count: 29
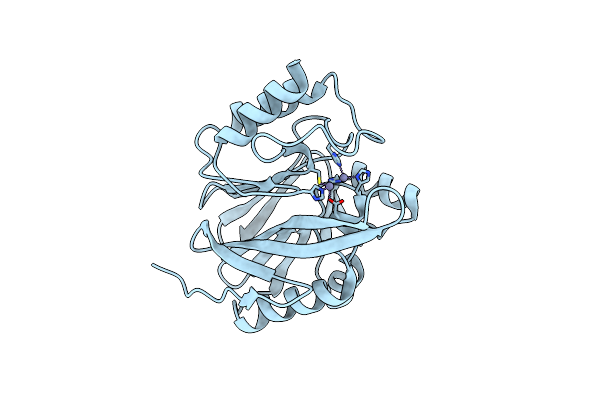 |
Crystal Structure Of Metallo-Beta-Lactamase Superfamily Protein Ccra-Like_Mbl-B1 From Dyadobacter Fermentans
Organism: Dyadobacter fermentans dsm 18053
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-07-17 Classification: HYDROLASE Ligands: ZN |
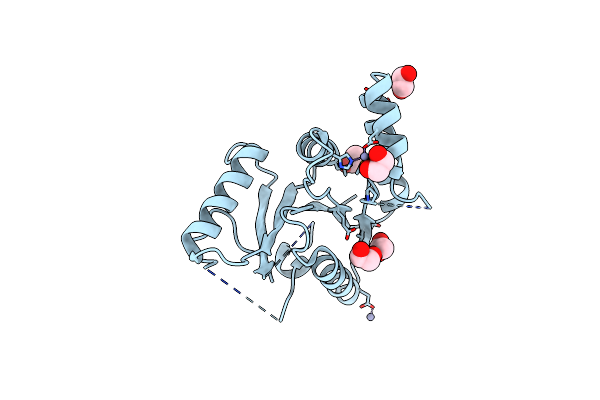 |
Crystal Structure Of The N-Terminal Domain Of The Phosphate Acetyltransferase From Escherichia Coli
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-12-29 Classification: TRANSFERASE Ligands: ACY, EDO, ZN |
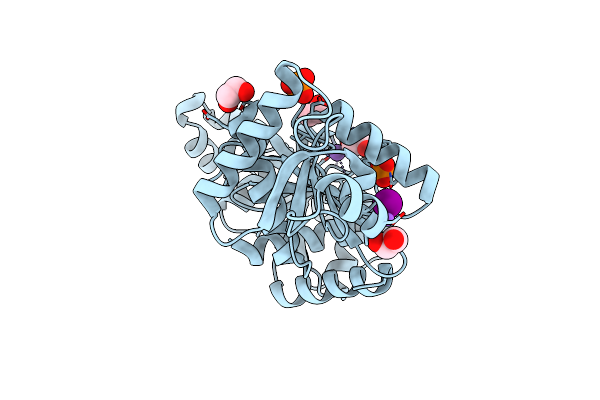 |
Crystal Structure Of The C-Terminal Domain Of The Phosphate Acetyltransferase From Escherichia Coli
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-12-22 Classification: TRANSFERASE Ligands: EDO, K, CL, PO4, IOD, NA |
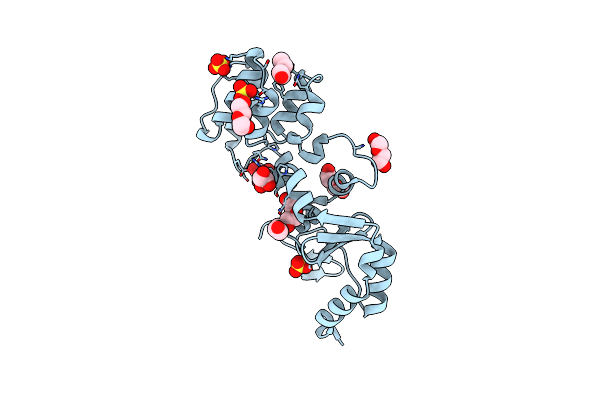 |
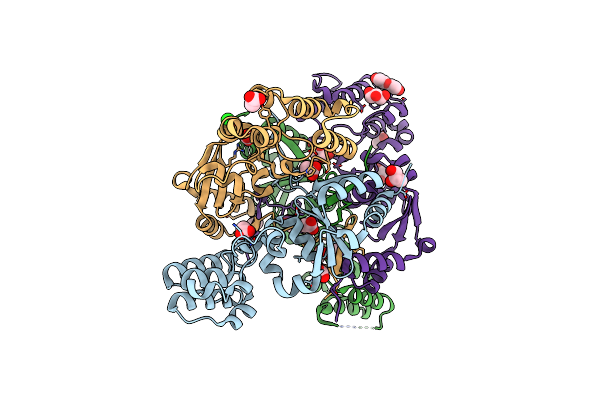
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, N239 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-01-27 Classification: SIGNALING PROTEIN Ligands: GOL, PEG, PG4, SO4 |
 |
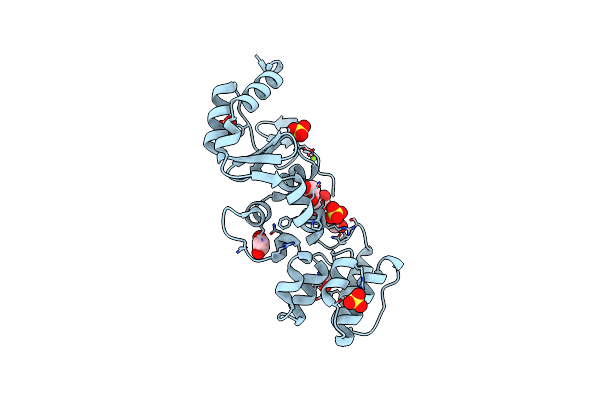
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, 2Nd Form
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: GOL, ACT, CL, PEG, SRT |
 |
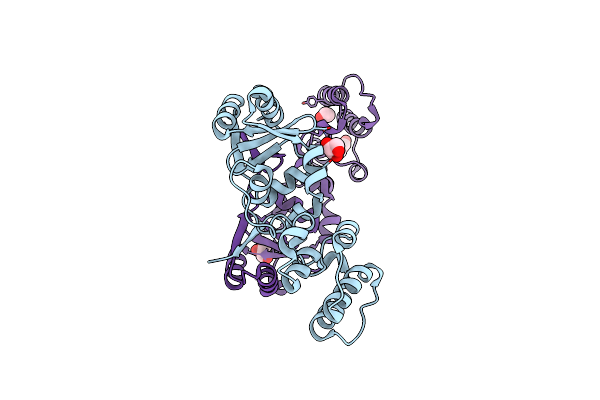
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, N239-T240 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: EDO, SO4, MG |
 |
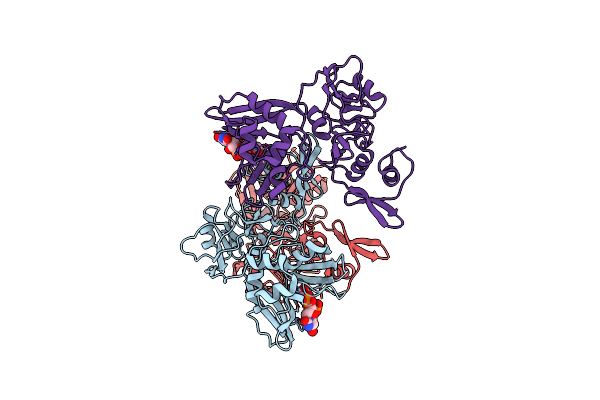
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, D238-T240 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: GOL, EDO |
 |
Crystal Structure Of Nsp15 Endoribonuclease From Sars Cov-2 In The Complex With Uridine-3',5'-Diphosphate
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-09-23 Classification: VIRAL PROTEIN Ligands: VQV, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-07-29 Classification: HYDROLASE Ligands: EDO, FMT |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: EDO |
 |
Crystal Structure Of Sialate O-Acetylesterase From Bacteroides Vulgatus By Serial Crystallography
Organism: Bacteroides vulgatus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-07-15 Classification: HYDROLASE |
 |
Crystal Structure Of Sialate O-Acetylesterase From Bacteroides Vulgatus With Microfluidics Crystals At Room Temperature
Organism: Bacteroides vulgatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-07-15 Classification: HYDROLASE |
 |
The Crystal Structure Of 3Cl Mainpro Of Sars-Cov-2 With Oxidized Cys145 (Sulfenic Acid Cysteine).
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: EDO, CL |
 |
The 1.28A Crystal Structure Of 3Cl Mainpro Of Sars-Cov-2 With Oxidized C145 (Sulfinic Acid Cysteine)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: EDO, FMT, ACT |
 |
Organism: Desulfarculus baarsii (strain atcc 33931 / dsm 2075 / vkm b-1802 / 2st14)
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2020-04-22 Classification: HYDROLASE Ligands: ALA, CIT |
 |
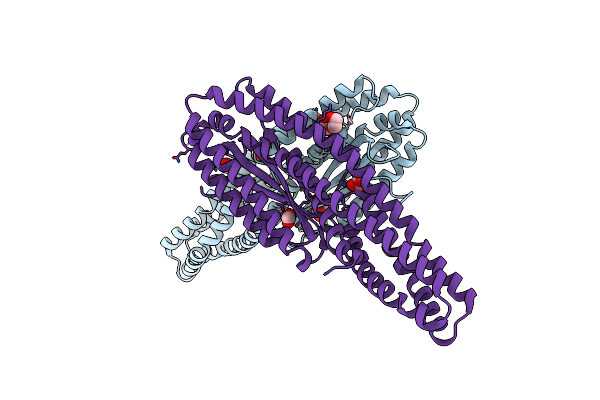
Crystal Structure Of Motility Associated Killing Factor B From Vibrio Cholerae
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2020-03-25 Classification: TOXIN Ligands: EDO |
 |
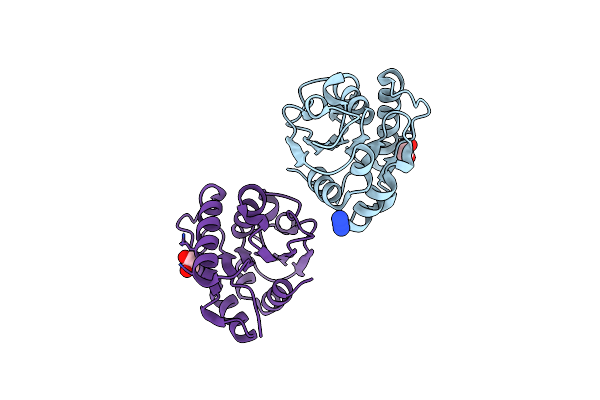
Crystal Structure Of Motility Associated Killing Factor E From Vibrio Cholerae
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-11 Classification: TOXIN Ligands: ACY, EDO, K, CL, FMT |
 |
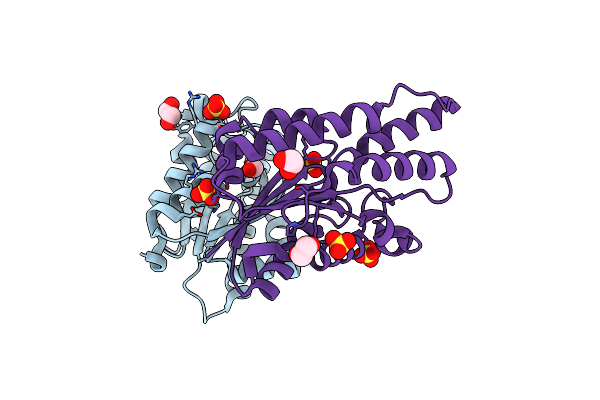
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2020-01-22 Classification: HYDROLASE Ligands: CL, EDO, AZI |
 |
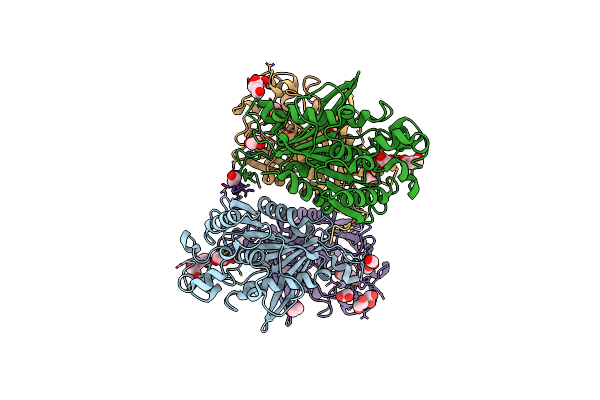
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-01-22 Classification: HYDROLASE Ligands: SO4, EDO, CL, FMT |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2020-01-22 Classification: HYDROLASE Ligands: EDO, PEG, SBT |

