Search Count: 24
 |
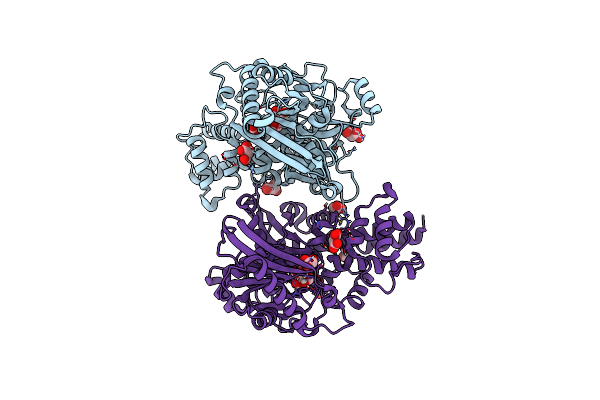
Crystal Structure Of Kluyveromyces Lactis Glucokinase In Complex With Glucose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: BGC, MLI |
 |
Crystal Structure Of Kluyveromyces Lactis Glucokinase In Complex With Mannose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MAN, MLI |
 |
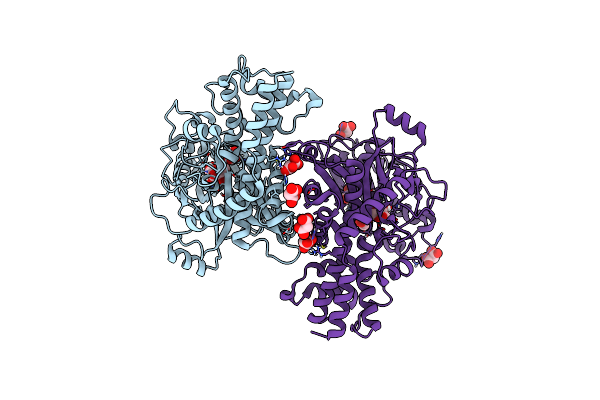
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Glucose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: BGC, MLI |
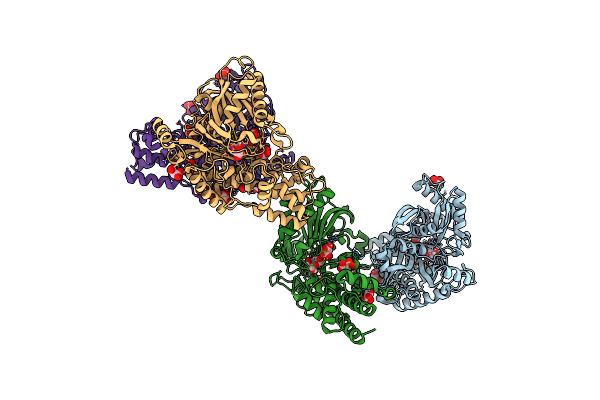 |
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Mannose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MAN, MLI |
 |
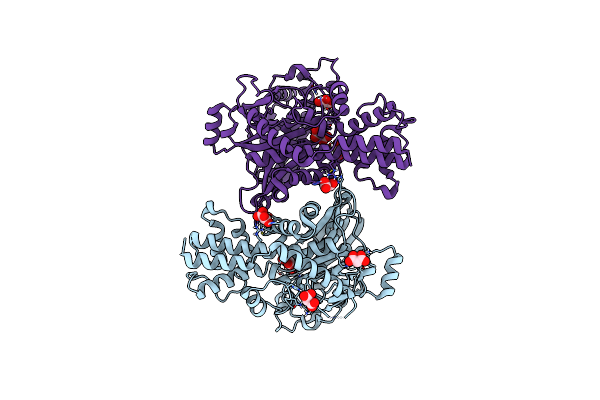
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Fructose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: FRU, MLI |
 |
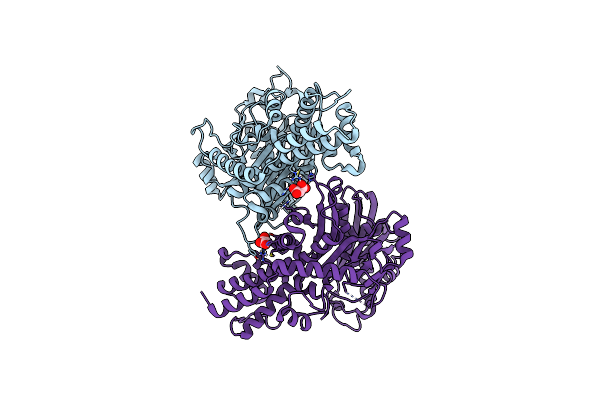
Crystal Structure Of The Intermediate Open State Of Apo Kluyveromyces Lactis Glucokinase
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MLI |
 |
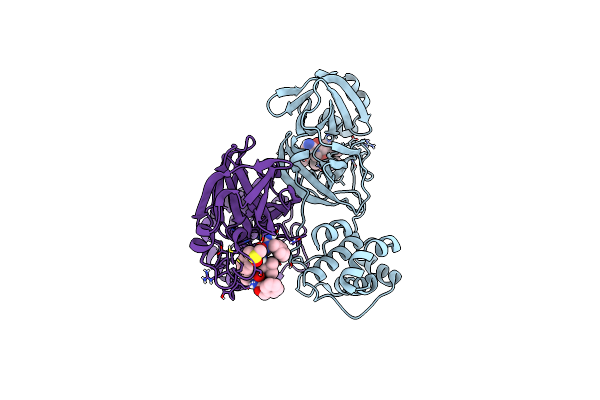
Crystal Structure Of The Open State Of Apo Kluyveromyces Lactis Glucokinase
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MLI |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Covalent Inhibitor Gue-3642 (Compound 1 In Publication)
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-07-03 Classification: VIRAL PROTEIN Ligands: XV9, DMS, CL |
 |
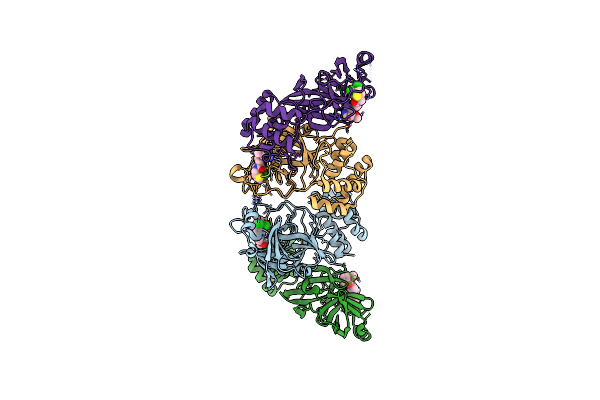
Crystal Structure Of Human Adenosine A2A Receptor (Construct A2A-Psb2-Bril) Complexed With The Partial Antagonist Luf5834 At The Orthosteric Pocket
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: CLR, OLA, OLC, OLB, PEG, A1H1S |
 |
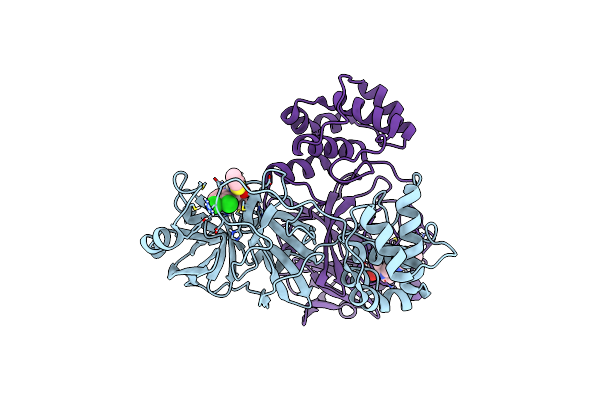
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Inhibitor Gc-67
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2023-12-06 Classification: VIRAL PROTEIN Ligands: KKO |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Inhibitor Gd-9
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: OZI, CL, BR |
 |
Crystal Structure Of The Adenosine A2A Receptor (Construct A2A-Psb2-Bril) Complexed With Etrumadenant At The Orthosteric Pocket
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-07-12 Classification: MEMBRANE PROTEIN Ligands: U30, CLR, OLA, OLC, OLB, PEG |
 |
Crystal Structure Of Stabilized A2A Adenosine Receptor A2Ar-Star2-Bril In Complex With Clinical Candidate Etrumadenant
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-05-31 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Variovorax Paradoxus Indole Monooxygenase (Vpinda1) In Complex With Fad
Organism: Variovorax paradoxus eps
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Variovorax Paradoxus Indole Monooxygenase (Vpinda1) In Complex With Indole
Organism: Variovorax paradoxus eps
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, IND, SO4, DMS, NA |
 |
Crystal Structure Of The F191M Variant Of Variovorax Paradoxus Indole Monooxygenase (Vpinda1) In Complex With 6-Bromoindole
Organism: Variovorax paradoxus eps
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, EDO, FK1 |
 |
Crystal Structure Of F191M Variant Variovorax Paradoxus Indole Monooxygenase (Vpinda1) In Complex With Methyl Phenyl Sulfide
Organism: Variovorax paradoxus eps
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, EDO, 16R |
 |
Crystal Structure Of F191M Variant Of Variovorax Paradoxus Indole Monooxygenase (Vpinda1) In Complex With Methyl Phenyl Sulfoxide
Organism: Variovorax paradoxus eps
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, IH0 |
 |
Crystal Structure Of The F191M/F201A Variant Of Variovorax Paradoxus Indole Monooxygenase (Vpinda1) In Complex With Benzyl Phenyl Sulfoxide
Organism: Variovorax paradoxus eps
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: IKF, FAD, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2022-10-19 Classification: SIGNALING PROTEIN Ligands: MG |

