Search Count: 16
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of The E.Coli 70S Ribosome In Complex With The Antibiotic Myxovalargin B.
Organism: Myxococcus fulvus, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: RIBOSOME Ligands: MG, ZN, FME, SPD |
 |
Cryo-Em Structure Of The E.Coli 50S Ribosomal Subunit In Complex With The Antibiotic Myxovalargin A.
Organism: Myxococcus fulvus, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-10-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-10-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: Viral Protein/Immune System Ligands: NAG |
 |
Circular Permutation Provides An Evolutionary Link Between Two Families Of Calcium-Dependent Carbohydrate Binding Modules
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-07-21 Classification: SUGAR BINDING PROTEIN Ligands: LMR, CA |
 |
Circular Permutation Provides An Evolutionary Link Between Two Families Of Calcium-Dependent Carbohydrate Binding Modules. Semet Form Of Vcbm60.
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2010-07-21 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
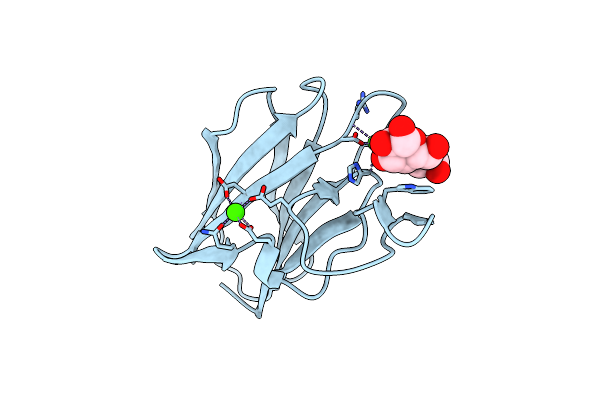
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2010-06-16 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2010-06-16 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-06-17 Classification: HYDROLASE Ligands: GOL |
 |
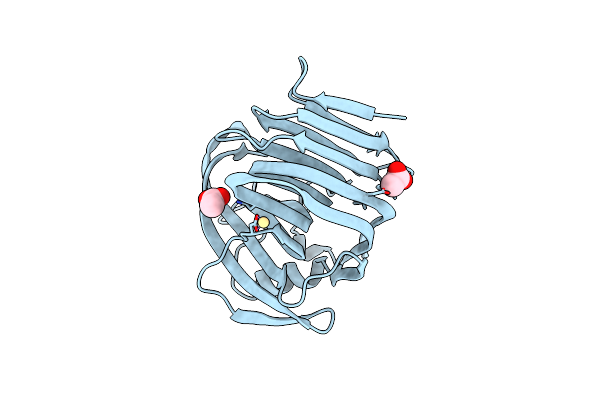
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-06-17 Classification: HYDROLASE Ligands: SO4, 12P |
 |
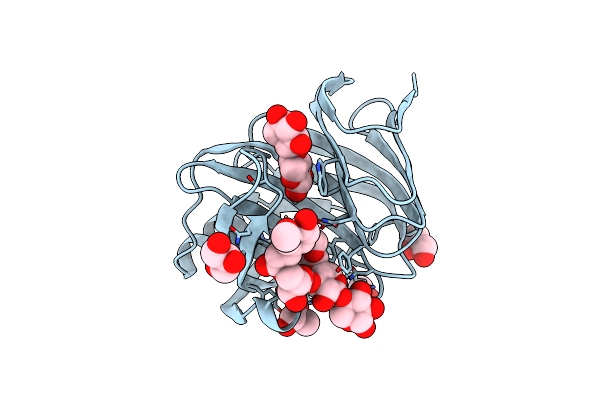
Organism: Neocallimastix patriciarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-12-25 Classification: HYDROLASE Ligands: CD, ACT |
 |
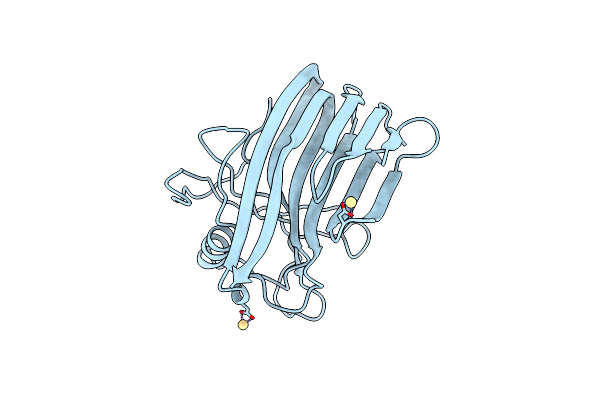
Crystal Structure Of Environmental Isolated Gh11 In Complex With Xylobiose And Feruloyl-Arabino-Xylotriose
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-12-25 Classification: HYDROLASE |
 |
Organism: Neocallimastix patriciarum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-02-13 Classification: HYDROLASE Ligands: CD |

