Search Count: 127
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
 |
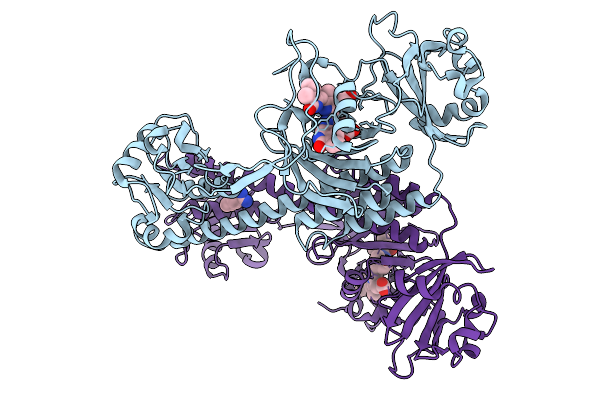
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
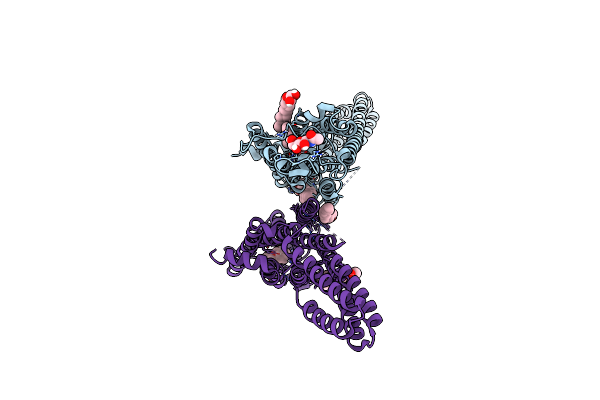
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
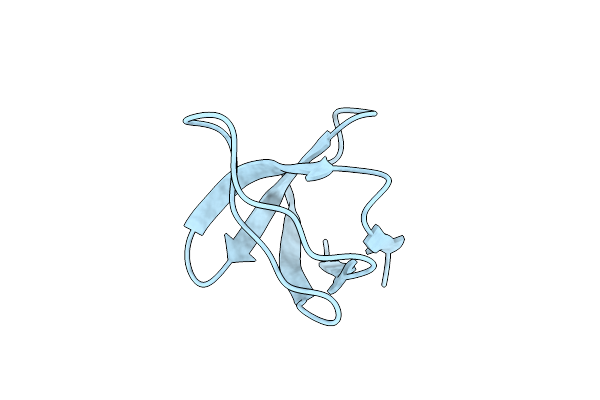
Xfel Crystal Structure Of The Human Sphingosine 1 Phosphate Receptor 5 In Complex With Ono-5430608
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-10 Classification: MEMBRANE PROTEIN Ligands: NAG, B2O, OLC, OLA |
 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Alpha Spectrin-Sh3 Domain
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-27 Classification: STRUCTURAL PROTEIN |
 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-27 Classification: HYDROLASE Ligands: CL |
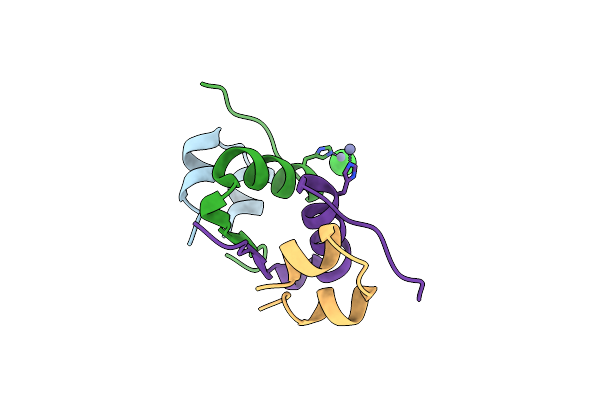 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Insulin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-10-27 Classification: HORMONE Ligands: ZN, CL |
 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Proteinase K
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-27 Classification: HYDROLASE Ligands: NO3, CA |
 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Phycocyanin
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-27 Classification: ELECTRON TRANSPORT Ligands: CYC, NA, CL |
 |
Xfel Crystal Structure Of The Prostaglandin D2 Receptor Crth2 In Complex With 15R-Methyl-Pgd2
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2021-08-25 Classification: MEMBRANE PROTEIN Ligands: YSS, NA, FLC, SO4 |
 |
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 50 Ps After Light Activation
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 100 Ps After Light Activation (0.90Mj/Mm2)
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 100 Ps After Light Activation (0.17Mj/Mm2)
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 100 Ps After Light Activation (2.63Mj/Mm2)
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 100 Ps After Light Activation (6.49 Mj/Mm2)
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
 |
2.0A Angstrom A2A Adenosine Receptor Structure Using Xfel Data Collected In Helium Atmosphere.
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-18 Classification: MEMBRANE PROTEIN |
 |
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-09-30 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |

