Search Count: 224
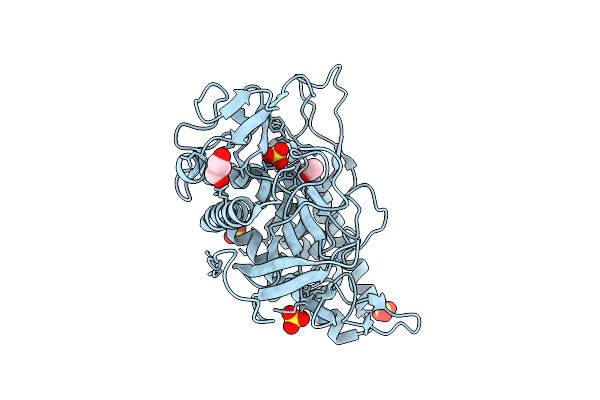 |
Crystal Structure Of The Catalytic Domain From The Bont-Like Toxin Complex Pg1 Of Paeniclostridium Ghonii
Organism: Paraclostridium ghonii
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TOXIN Ligands: ZN, SO4, ACT, GOL |
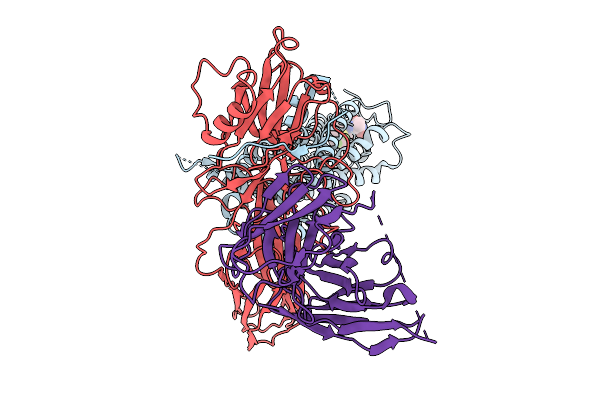 |
Cryo-Em Structure Of The Orphan Gpr52 Beta-Arrestin 1 Complex Bound To Ligand C17
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: EN6 |
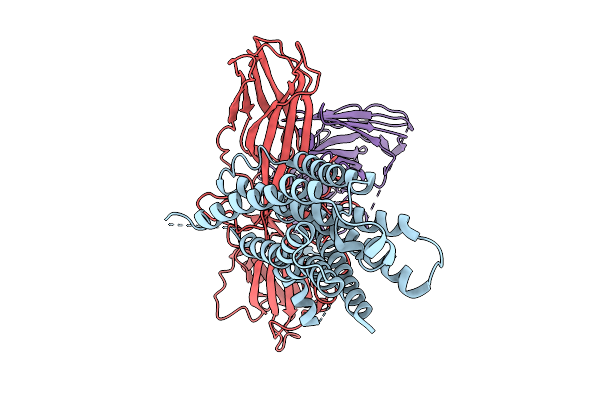 |
Cryo-Em Structure Of The Orphan Gpr52 Bound To Beta-Arrestin 1 In Ligand-Free State
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
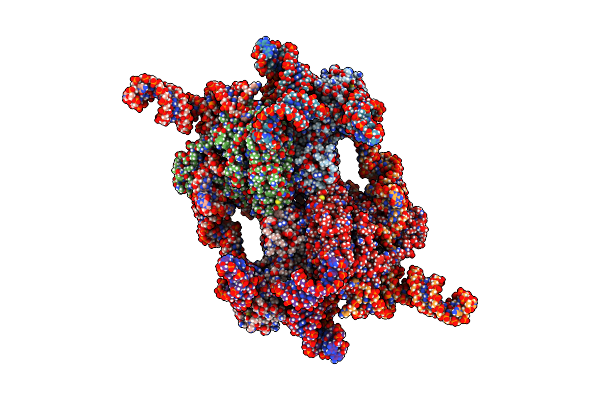 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: ANTIVIRAL PROTEIN/RNA |
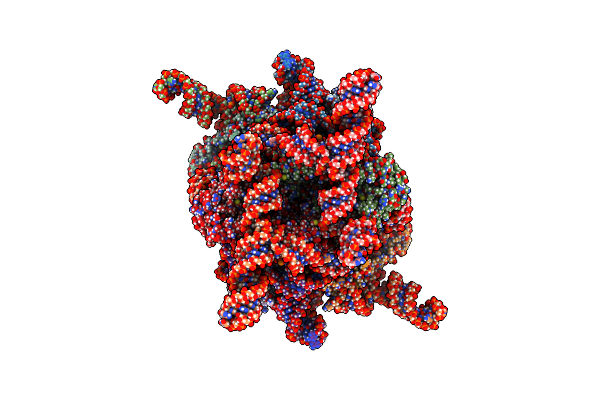 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: ANTIVIRAL PROTEIN/RNA |
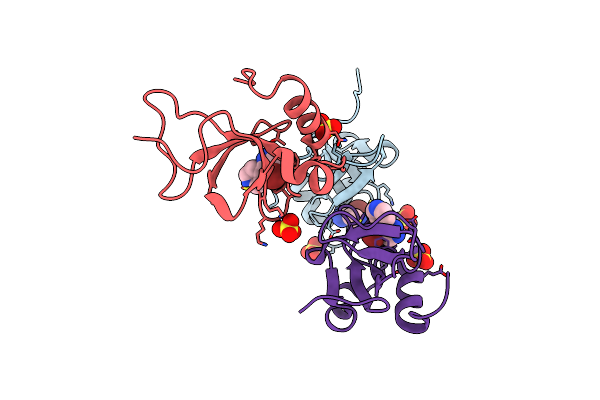 |
Crystal Structure Of Hdgfrp2 Pwwp Domain In Complex With 4-(4-Bromo-1H-Pyrazol-3-Yl) Pyridine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: SIGNALING PROTEIN Ligands: A1D73, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: SIGNALING PROTEIN Ligands: DMS, QMR |
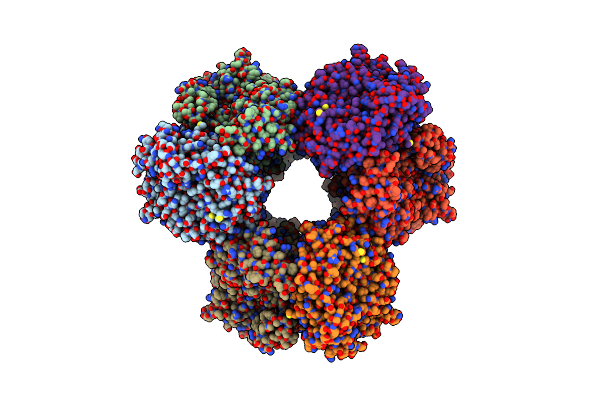 |
Enhancing The Synthesis Efficiency Of Galacto-Oligosaccharides Of A Beta-Galactosidase From Paenibacillus Barengoltzii By Engineering The Active And Distal Sites
Organism: Paenibacillus barengoltzii
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2025-04-02 Classification: TRANSFERASE |
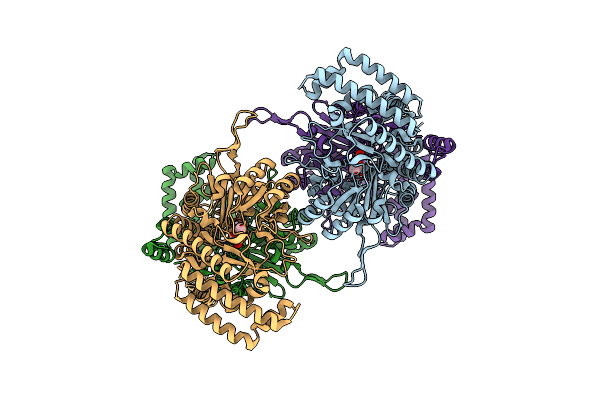 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GNP, A1AOV, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GNP, MG, BTB, A1AOM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GNP, MG, A1AOD, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GNP, MG, A1AOK |
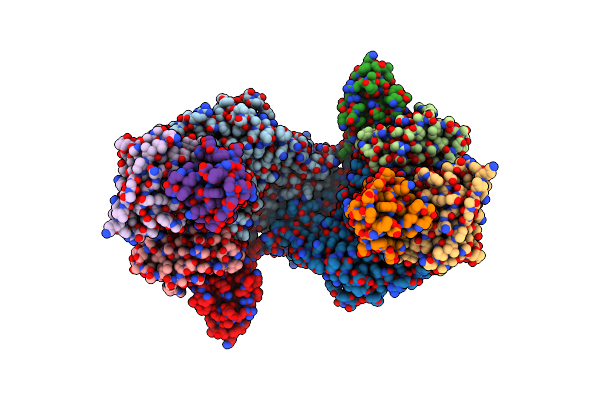 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: IMMUNE SYSTEM Ligands: ZN, NA, CA |
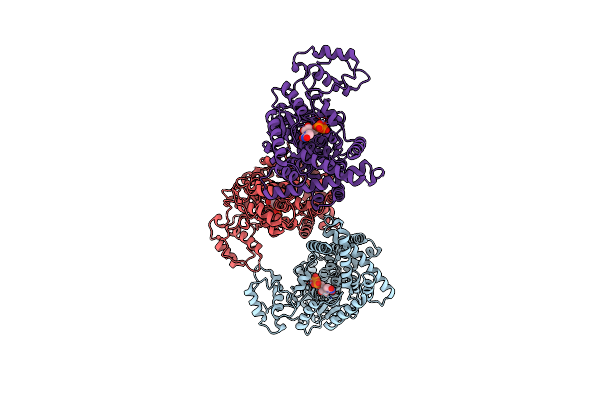 |
The Crystal Structure Of Glucosyltransferase Tcdb From Clostridioides Difficile
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:3.95 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: MN, UDP |
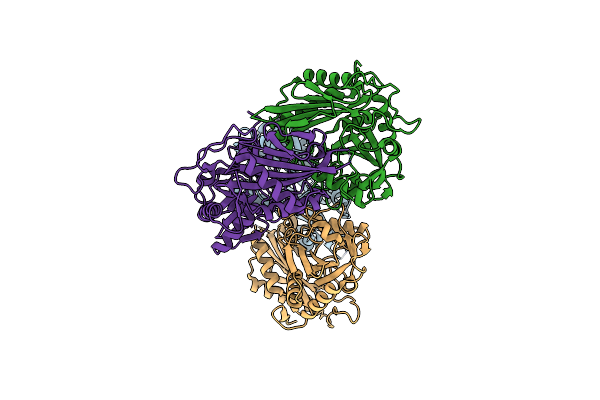 |
Structure Of Wild-Type Aminotransferase From Mycolicibacterium Neoaurum In Complex With Llp
Organism: Mycolicibacterium neoaurum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-25 Classification: TRANSFERASE |
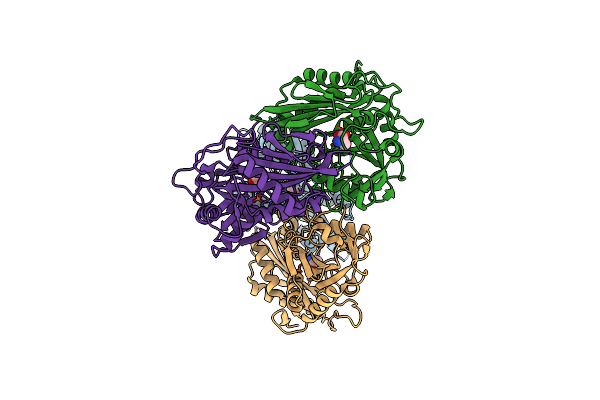 |
Structure Of Wild-Type Aminotransferase From Mycolicibacterium Neoaurum In Complex With Llp And Ala
Organism: Mycolicibacterium neoaurum vkm ac-1815d
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: ALA |
 |
Organism: Geobacter sulfurreducens, Escherichia coli 'bl21-gold(de3)plyss ag'
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: STRUCTURAL PROTEIN/RNA/DNA Ligands: MG |
 |
Organism: Geobacter sulfurreducens, Escherichia coli 'bl21-gold(de3)plyss ag'
Method: ELECTRON MICROSCOPY Resolution:3.38 Å Release Date: 2024-10-16 Classification: STRUCTURAL PROTEIN/RNA/DNA Ligands: MG |

