Search Count: 94
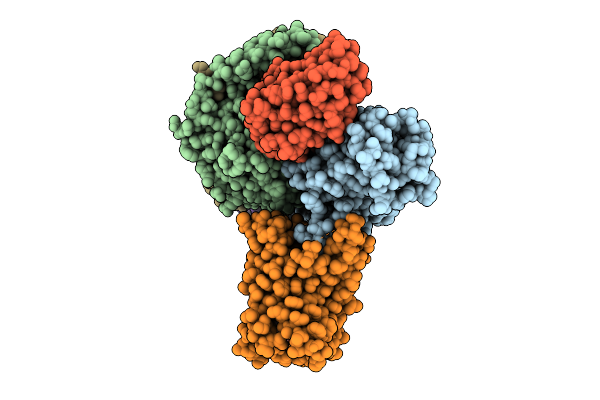 |
Organism: Spodoptera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: AND |
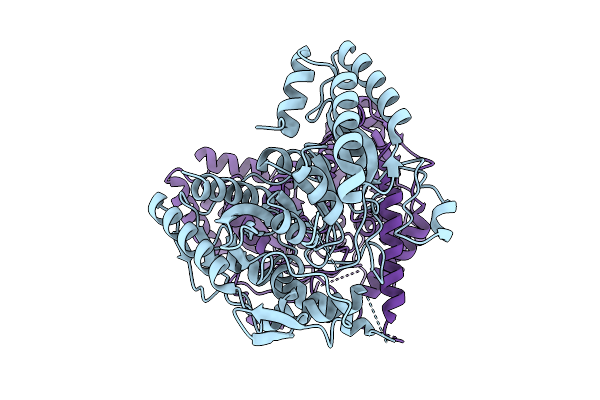 |
Organism: Sphingomonas sp. tpd3009
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: HYDROLASE Ligands: GOL, SO4 |
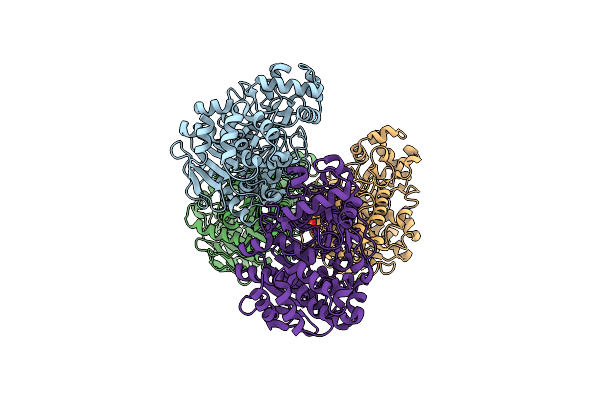 |
Crystal Structure Of A Novel Pu Plastic Degradation Enzyme From Thermaerobacter Marianensis
Organism: Thermaerobacter marianensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: SO4 |
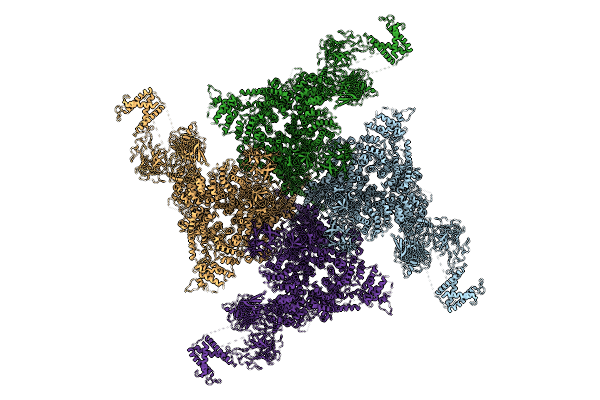 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: CA, ZN |
 |
Cryo-Em Structure Of Ryanodine Receptor 1 (Tm Helix S0,100 Nm Ca2+, Closed State)
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of Ryanodine Receptor 1 (Tm Helix S0,100 Nm Ca2+, Open State)
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: ATP, CA |
 |
Cryo-Em Structure Of Ryanodine Receptor 1 (Tm Helix S0,20 Um Ca2+, 2 Mm Atp)
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: CA, ATP |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: ZN, CA |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: PG6, SO4 |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: SO4, PMS |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: ACT, A1IHK |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Enzyme With Ligand From Thermaerobacter Marianensis
Organism: Thermaerobacter marianensis (strain atcc 700841 / dsm 12885 / jcm 10246 / 7p75a)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: SO4, A1D5F |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 From Uncultured Bacterium In Complex With Ligand
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: A1D5H |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 Mutant From Uncultured Bacterium In Complex With Ligand
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: A1D5H, GOL |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Enzyme From Thermaerobacter Marianensis
Organism: Thermaerobacter marianensis (strain atcc 700841 / dsm 12885 / jcm 10246 / 7p75a)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-18 Classification: HYDROLASE |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 From Uncultured Bacterium
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: GOL, SO4, PMS |
 |
Urethanase Umg-Sp1 Without Inhibitor Or Substrate Displays Flexible Active Site Loops
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2024-07-24 Classification: HYDROLASE |

