Search Count: 15
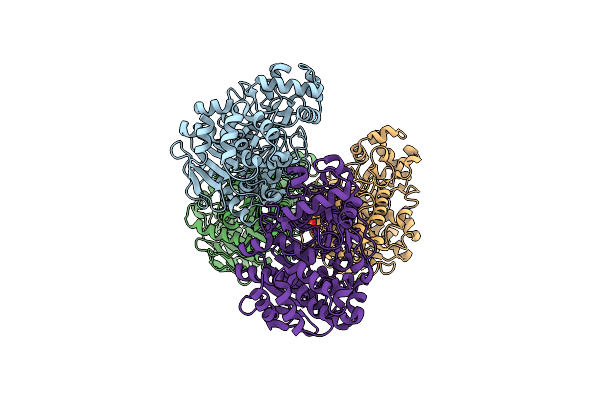 |
Crystal Structure Of A Novel Pu Plastic Degradation Enzyme From Thermaerobacter Marianensis
Organism: Thermaerobacter marianensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: SO4 |
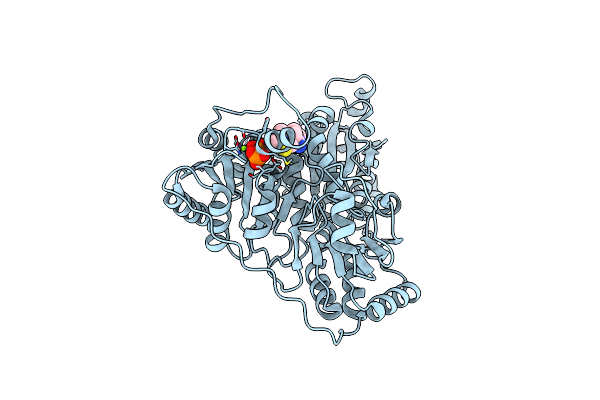 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, MG |
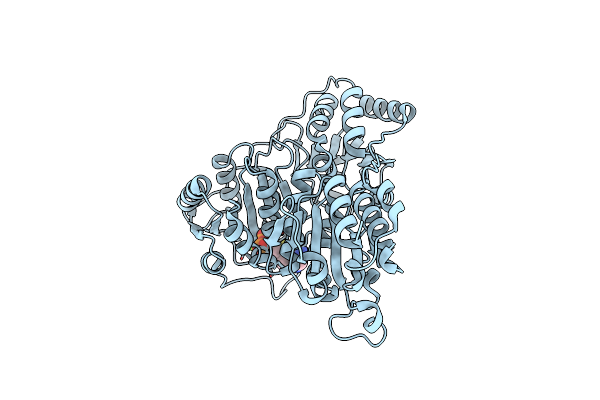 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, MG |
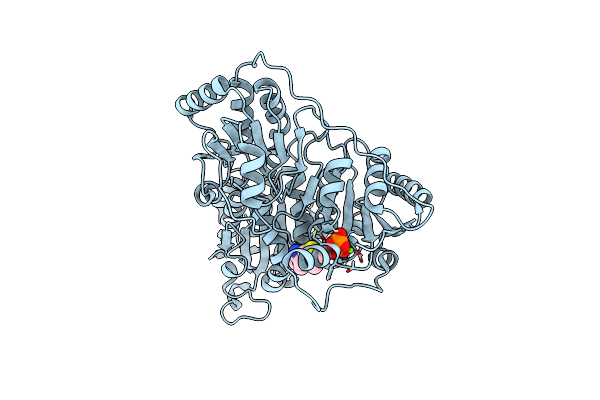 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, MG |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Enzyme From Thermaerobacter Marianensis
Organism: Thermaerobacter marianensis (strain atcc 700841 / dsm 12885 / jcm 10246 / 7p75a)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-18 Classification: HYDROLASE |
 |
Crystal Structure Of A Novel Ketoreductase From Sphingobacterium Siyangense Sy1
Organism: Sphingobacterium siyangense
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of A Ketoreductase From Sphingobacterium Siyangense Sy1 With Co-Enzyme
Organism: Sphingobacterium siyangense
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Hypoxylon sp. e7406b
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-02-28 Classification: TRANSFERASE |
 |
Organism: Hypoxylon sp. e7406b
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2024-02-28 Classification: TRANSFERASE |
 |
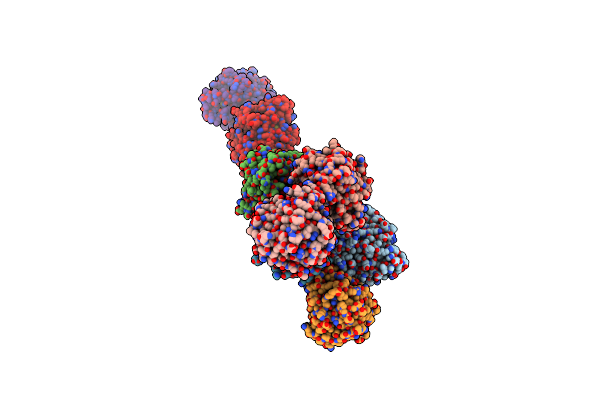
Organism: Rhinocladiella mackenziei cbs 650.93
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2018-05-02 Classification: HYDROLASE Ligands: 1PE |
 |
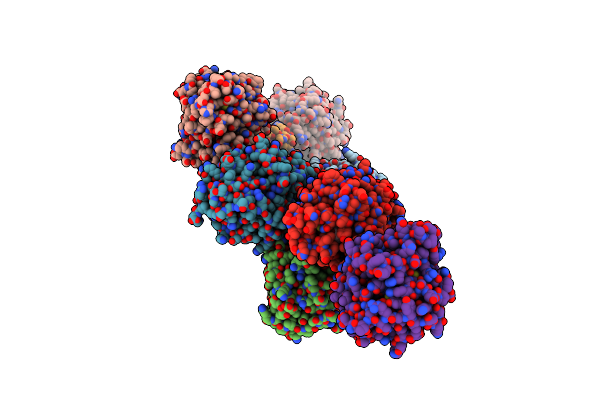
Organism: Rhinocladiella mackenziei cbs 650.93
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-05-02 Classification: HYDROLASE Ligands: 36J |
 |
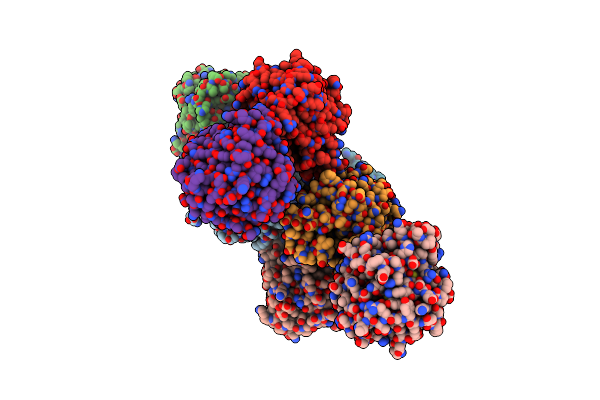
Organism: Rhinocladiella mackenziei cbs 650.93
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-05-02 Classification: HYDROLASE Ligands: ZER |
 |
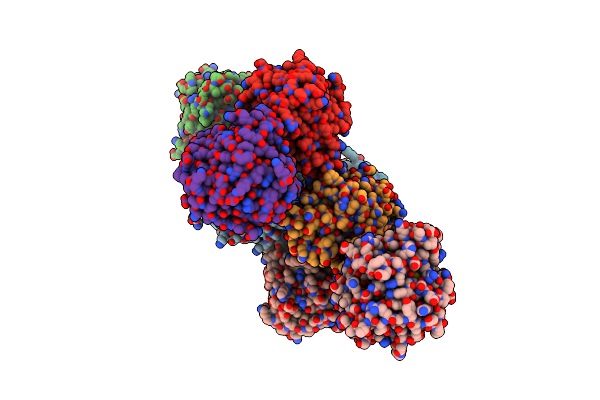
Organism: Rhinocladiella mackenziei cbs 650.93
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-05-02 Classification: HYDROLASE Ligands: PEG |
 |
Organism: Rhinocladiella mackenziei cbs 650.93
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-05-02 Classification: HYDROLASE Ligands: 36J, PEG |
 |
Organism: Rhinocladiella mackenziei cbs 650.93
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2018-05-02 Classification: HYDROLASE Ligands: ZER, SO4 |

