Search Count: 218
 |
Crystal Structure Of An Agose Isomerase Mutant 5 (Tst4Ease M5) From Thermotogota Bacterium With Zn
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: ISOMERASE Ligands: ZN |
 |
Crystal Structure Of A Wild-Type Tagose Isomerase (Tst4Ease Wt) From Thermotogota Bacterium Complex With Zn
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: ISOMERASE Ligands: ZN |
 |
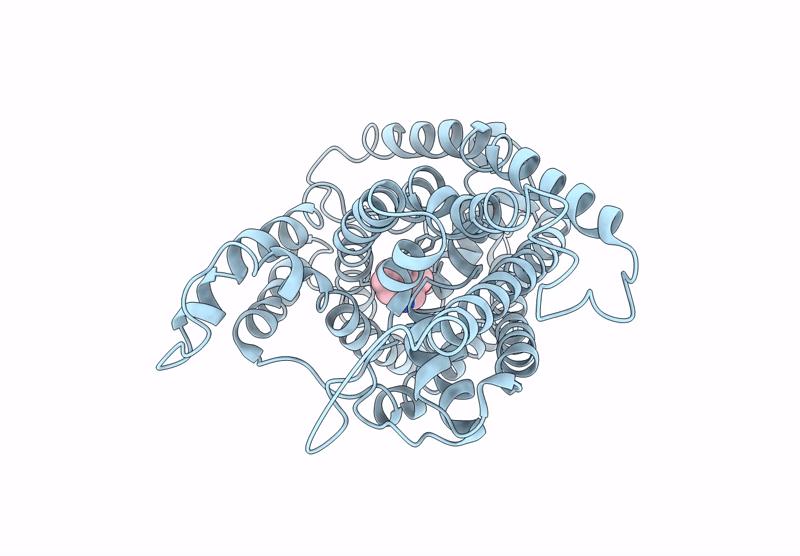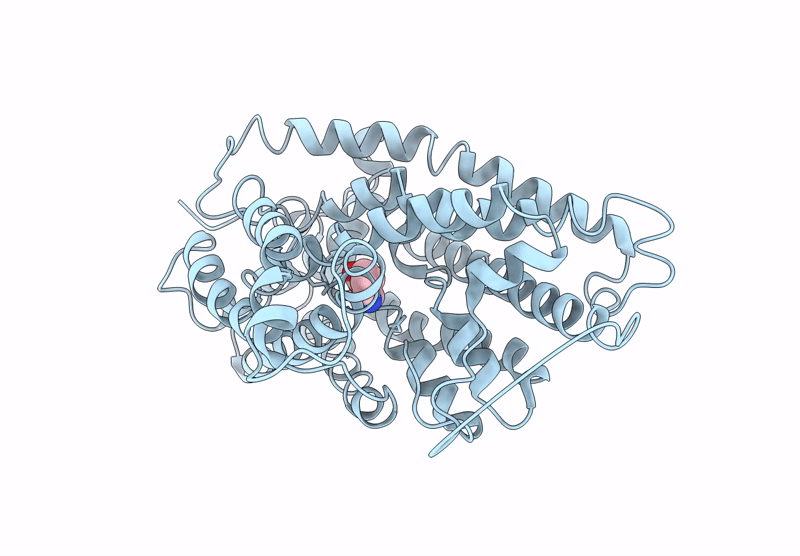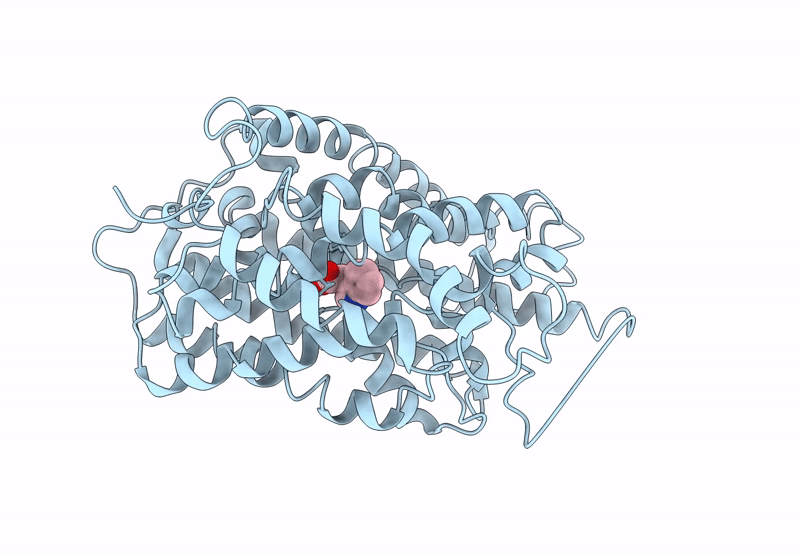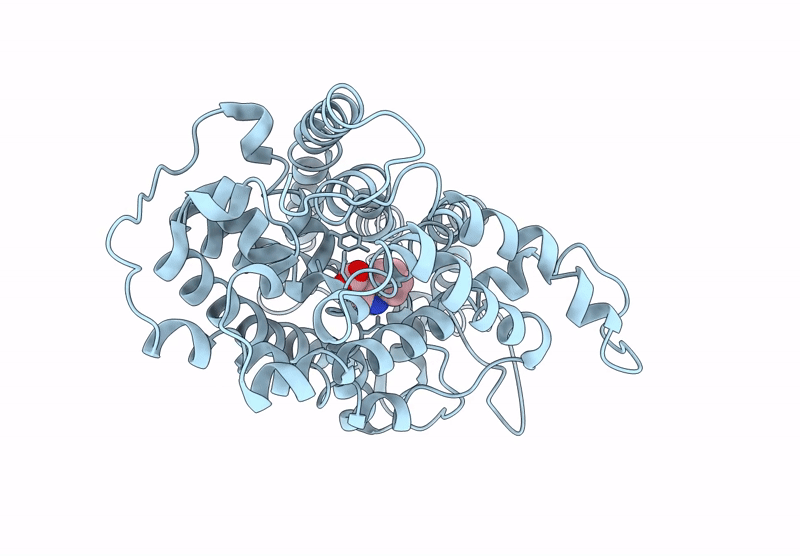
Structure Of Aminotransferase From Mycolicibacterium Neoaurum In Complex With Plp
Organism: Mycolicibacterium neoaurum
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: TRANSFERASE Ligands: PLP |
 |
Structure Of The Auxin Importer Aux1 In Arabidopsis Thaliana In The Iaa-Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN Ligands: IAC |
 |
Structure Of The Auxin Importer Aux1 In Arabidopsis Thaliana In The Apo State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Auxin Importer Aux1 In Arabidopsis Thaliana In The Chpaa-Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: MEMBRANE PROTEIN Ligands: 3C4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2025-05-21 Classification: APOPTOSIS |
 |
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Apo State At Ph 5.5
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN |
 |
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Apo State At Ph 7.4
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN |
 |
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Trans-Zeatin-Bound State At Ph 5.5
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN Ligands: ZEA |
 |
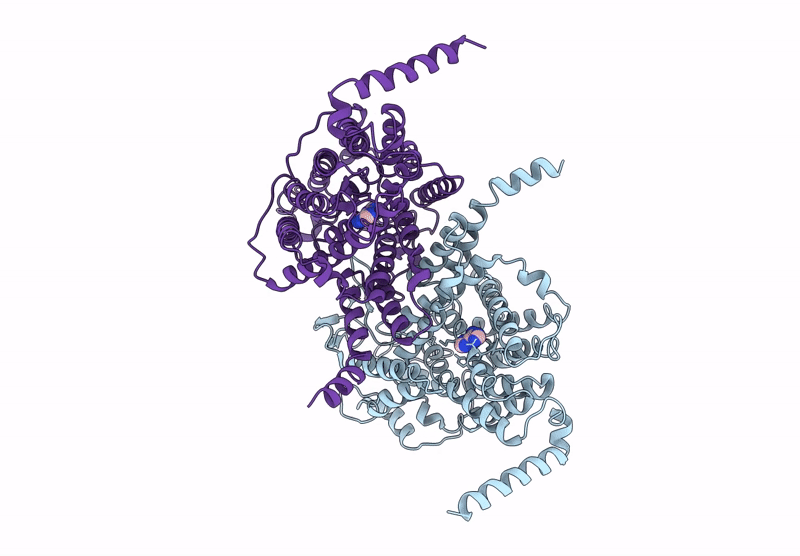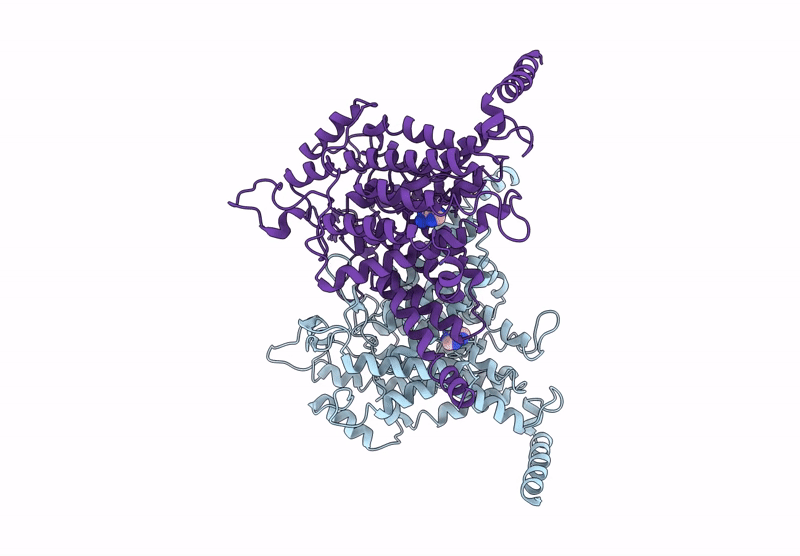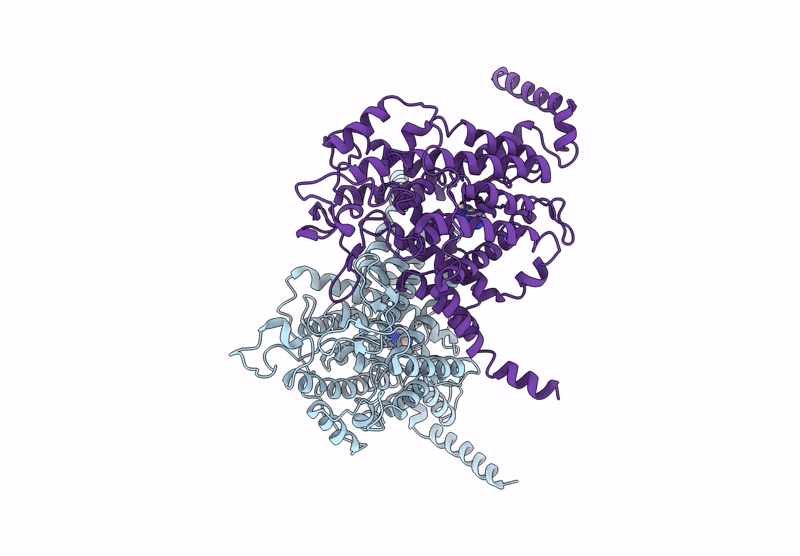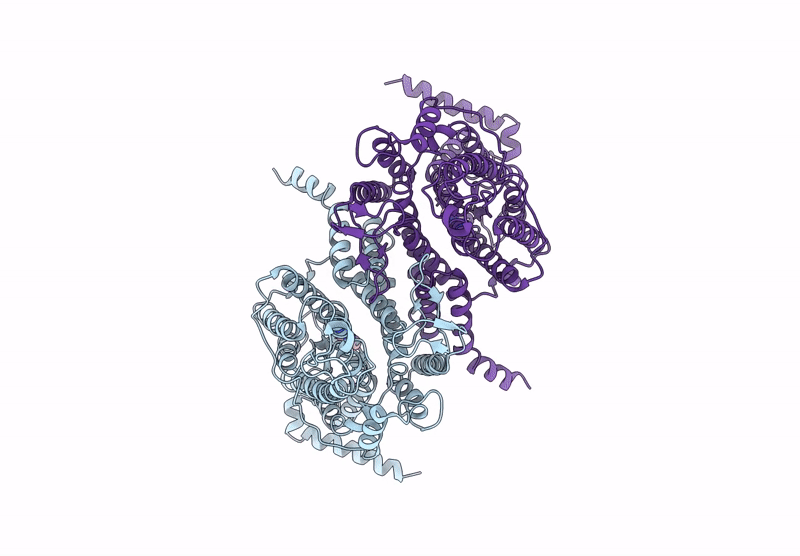
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Adenine-Bound State At Ph 5.5
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN Ligands: ADE |
 |
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Adenine-Bound State At Ph 7.4
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN Ligands: ADE |
 |
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Trans-Zeatin-Bound State-1 At Ph 7.4
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN Ligands: ZEA |
 |
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Trans-Zeatin-Bound State-2 At Ph 7.4
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN Ligands: ZEA |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Enzyme From Thermaerobacter Marianensis
Organism: Thermaerobacter marianensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: TRANSPORT PROTEIN |
 |
Structure Of The Wild-Type Arabidopsis Abcb1 In The Brassinolide-Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: TRANSPORT PROTEIN Ligands: BLD |
 |
Structure Of The Arabidopsis Abcb1 Eq Mutant In The Bl Plus Atp Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: TRANSPORT PROTEIN Ligands: ATP, BLD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: APOPTOSIS Ligands: ZN |
 |

