Search Count: 45
 |
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: MG, JHN |
 |
Structure Of Mycobacterium Tuberculosis Steb (Rv1698), A Cell Division Regulator
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: CELL CYCLE Ligands: CD, ACY, PG4 |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: CELL CYCLE Ligands: HEZ |
 |
Structure Of Mycobacterium Tuberculosis Stea (Rv1697), A Cell Division Regulator
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: CELL CYCLE |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: CELL CYCLE |
 |
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: DNA BINDING PROTEIN Ligands: A1H5Q |
 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: STRUCTURAL PROTEIN |
 |
Structure Of The Catalytic Domain Of P. Vivax Sub1 (Triclinic Crystal Form) In Complex With Inhibitor
Organism: Plasmodium vivax, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: NAG, CA, SO4 |
 |
Structure Of The Catalytic Domain Of P. Vivax Sub1 (Triclinic Crystal Form)
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: CA, NAG, SO4 |
 |
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: CA |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2023-03-29 Classification: METAL BINDING PROTEIN |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-02-22 Classification: CELL CYCLE |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-02-22 Classification: CELL CYCLE |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-01-18 Classification: CELL CYCLE Ligands: CA |
 |
Structure Of Peptidoglycan Hydrolase Cg1735 From Corynebacterium Glutamicum, Trigonal Crystal Form
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-01-18 Classification: CELL CYCLE Ligands: PC4 |
 |
Structure Of Peptidoglycan Hydrolase Cg1735 From Corynebacterium Glutamicum, Orthorhombic Crystal Form
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2023-01-18 Classification: CELL CYCLE |
 |
Organism: Methanobrevibacter smithii (strain atcc 35061 / dsm 861 / ocm 144 / ps)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-03-31 Classification: CELL CYCLE Ligands: TRS |
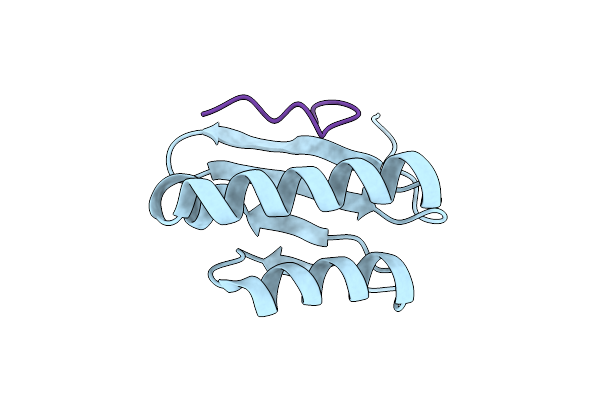 |
Cell Division Protein Sepf From Methanobrevibacter Smithii In Complex With Ftsz-Ctd
Organism: Methanobrevibacter smithii (strain atcc 35061 / dsm 861 / ocm 144 / ps), Methanobrevibacter smithii dsm 2375
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-03-31 Classification: CELL CYCLE |
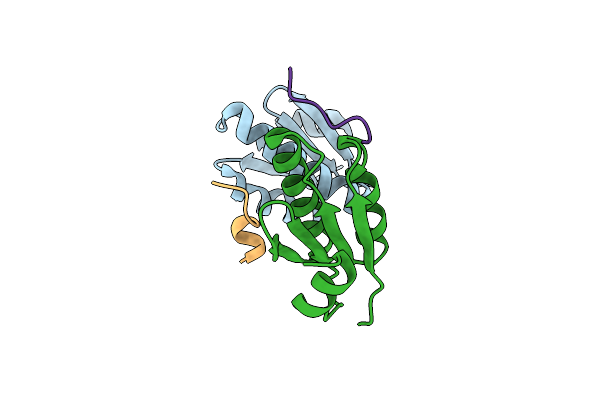 |
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-03-11 Classification: CELL CYCLE |
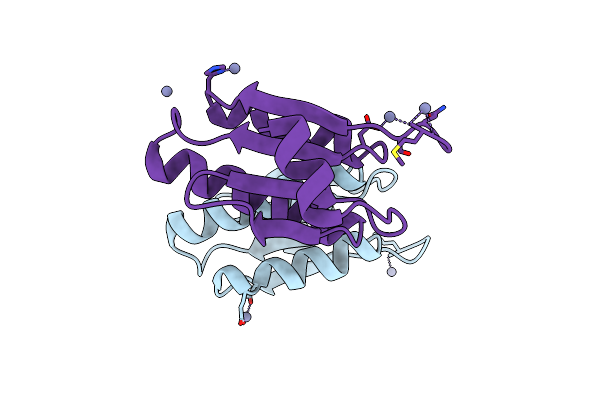 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-03-11 Classification: CELL CYCLE Ligands: ZN |

