Search Count: 20
 |
The Crystal Structure Of The Antimycin Pathway Standalone Ketoreductase, Antm
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-06-30 Classification: OXIDOREDUCTASE |
 |
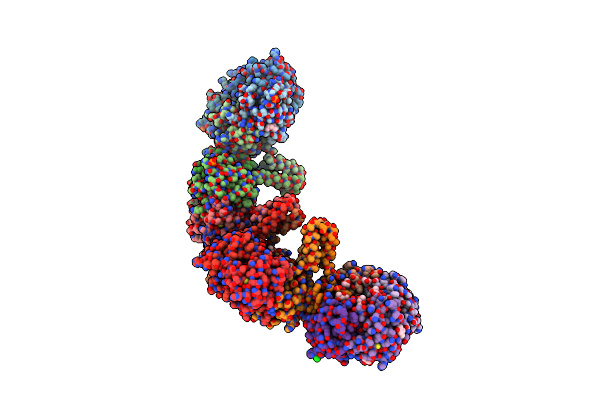
The Crystal Structure Of The Antimycin Pathway Standalone Ketoreductase, Antm, In Complex With Nadph
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-06-30 Classification: OXIDOREDUCTASE Ligands: NDP, GOL |
 |
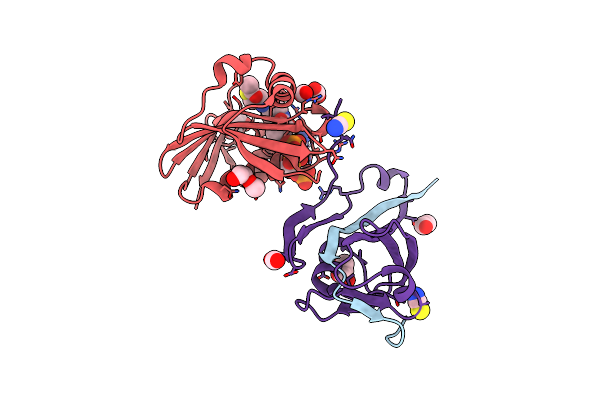
Engineered Higher-Order Assembly Of Cholera Toxin B Subunits Via The Addition Of C-Terminal Parallel Coiled-Coiled Domains
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-03-20 Classification: TOXIN Ligands: PO4, MPD, CL |
 |
Complex Of Wild Type E. Coli Alpha Aspartate Decarboxylase With Its Processing Factor Panz
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2017-09-13 Classification: LYASE Ligands: GOL, PEG, ACO, MG, CO2, SCN, 74C |
 |
E. Coli L-Aspartate-Alpha-Decarboxylase Mutant N72Q To A Resolution Of 1.9 Angstroms
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-01-13 Classification: LYASE Ligands: PEG, SCN |
 |
Direct Visualisation Of Strain-Induced Protein Post-Translational Modification
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2015-03-25 Classification: LYASE Ligands: ACO, MG, CL |
 |
Direct Visualisation Of Strain-Induced Protein Prost-Translational Modification
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-03-25 Classification: LYASE Ligands: SCN, ACO, MG |
 |
Direct Visualisation Of Strain-Induced Protein Post-Translational Modification
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-03-25 Classification: LYASE Ligands: SCN, ACO, MG |
 |
Conformational Dynamics Of Aspartate Alpha-Decarboxylase Active Site Revealed By Protein-Ligand Complexes: 1-Methyl-L-Aspartate Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-10-31 Classification: HYDROLASE |
 |
Conformational Dynamics Of Aspartate Alpha-Decarboxylase Active Site Revealed By Protein-Ligand Complexes: 1-Methyl-L-Aspartate Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-10-31 Classification: HYDROLASE Ligands: GLU |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2012-07-25 Classification: LYASE Ligands: MLI |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-04-11 Classification: LYASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2003-11-18 Classification: LYASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2003-11-18 Classification: LYASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-11-18 Classification: LYASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2003-11-18 Classification: LYASE |
 |
Unprocessed Aspartate Decarboxylase Mutant, With Alanine Inserted At Position 24
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-11-18 Classification: LYASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-11-18 Classification: LYASE Ligands: SO4 |
 |
Unprocessed Pyruvoyl Dependent Aspartate Decarboxylase With An Alanine Insertion At Position 26
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-11-11 Classification: LYASE Ligands: SO4 |
 |
Unprocessed Pyruvoyl Dependent Aspartate Decarboxylase With Histidine 11 Mutated To Alanine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-11-11 Classification: LYASE Ligands: SO4 |

