Search Count: 58
 |
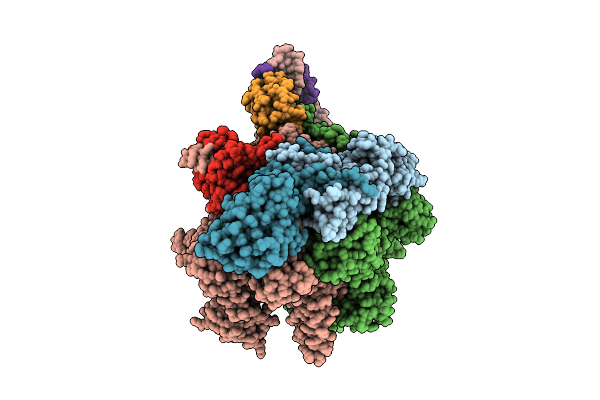
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex In The Presence Of Atp
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, ZN, 1N7 |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex In The Presence Of Ap4A
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: ZN, MG, 1N7 |
 |
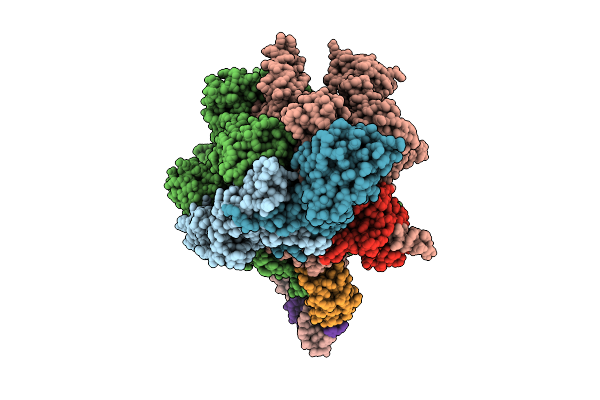
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Atp-C, -1 Dc In The Template Dna Strand
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, ZN, C5P, ATP |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex In The Presence Of Up4A
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, ZN |
 |
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: ZN, MG, 1N7 |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Ap4A-C, -1 Da In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, ZN, C5P, B4P |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Ap4A-C, -1 Dc In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: ZN, MG, C5P, B4P, 1N7 |
 |
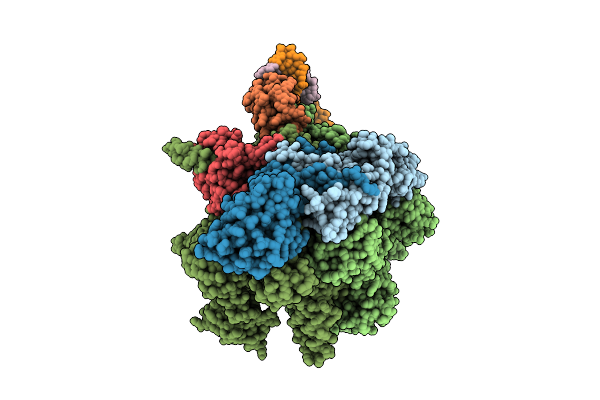
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex In The Presence Of Gp4A, -1 Da In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, ZN |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Gp4A-C-U And Ctp, -1 Dc In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, CTP, ZN |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Gp4A-C, -1 Dc In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: ZN, MG, C5P, 1N7, A1IF9 |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Up4A-C And Up4A, -1 Da In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, A1L89, ZN, C5P |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Up4A-C And Utp, -1 Da In The Template Dna Strand
Organism: Thermus thermophilus hb8, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, ZN, 1N7, C5P, UTP, A1L89 |
 |
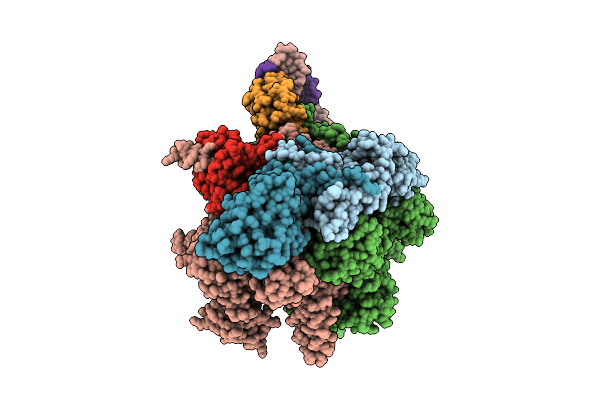
Cryoem Structure Of T.Thermophilus Transcription Initiation Complex Bound To Gp4A-C, -1 Da In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, A1IF9, C5P, ZN |
 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex In The Presence Of Gp4A, -1 Dc In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, A1L9G, ZN |
 |
Crystal Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Ap4G
Organism: Thermus thermophilus hb8, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, A1IF9, TRS, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-16 Classification: CHAPERONE Ligands: A1AG2, MG |
 |
Crystal Structure Of R. Sphaeroides Photosynthetic Reaction Center Variant Y(M210)2-Cyanophenylalanine
Organism: Cereibacter sphaeroides
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: PHOTOSYNTHESIS Ligands: BCL, BPH, U10, CL, LDA, FE, SPO, CDL |
 |
Crystal Structure Of R.Sphaeroides Photosynthetic Reaction Center Variant Y(M210)2-Chlorophenylalanine
Organism: Cereibacter sphaeroides
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: PHOTOSYNTHESIS Ligands: FE, BPH, BCL, U10, CL, LDA, SPO, CDL |
 |
Crystal Structure Of R. Sphaeroides Photosynthetic Reaction Center Variant Y(M210)2-Methoxyphenylalanine
Organism: Cereibacter sphaeroides
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: PHOTOSYNTHESIS Ligands: BPH, BCL, CL, LDA, FE, SPO, CDL |
 |
Crystal Structure Of R. Sphaeroides Photosynthetic Reaction Center Variant Y(M210)2-Bromophenylalanine
Organism: Cereibacter sphaeroides
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: PHOTOSYNTHESIS Ligands: LDA, BCL, CL, FE, BPH, SPO, CDL |

