Search Count: 17
 |
Organism: Escherichia coli ms 115-1
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-12-25 Classification: DNA BINDING PROTEIN Ligands: MG, P6G, PGE, CL |
 |
Organism: Escherichia coli ms 115-1
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-12-25 Classification: DNA BINDING PROTEIN Ligands: CL |
 |
Organism: Escherichia coli ms 115-1
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2019-12-25 Classification: DNA BINDING PROTEIN Ligands: CL |
 |
Organism: Escherichia coli ms 115-1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-25 Classification: PROTEIN BINDING Ligands: ATP, MG, CL, MPD |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-12-25 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2019-12-25 Classification: SIGNALING PROTEIN Ligands: ATP, CL, MG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-12-25 Classification: PROTEIN BINDING Ligands: NI, CL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-12-25 Classification: PROTEIN BINDING Ligands: CA |
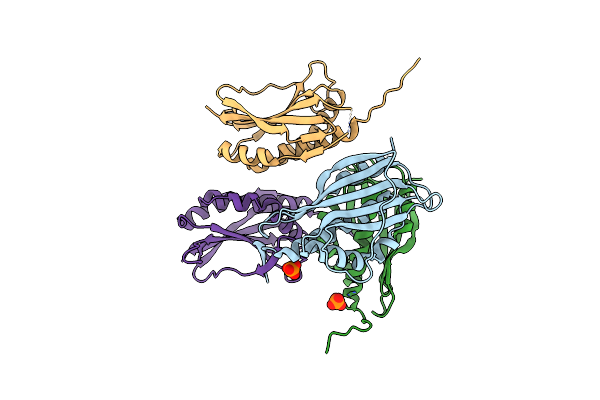 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2019-12-25 Classification: PROTEIN BINDING Ligands: PO4 |
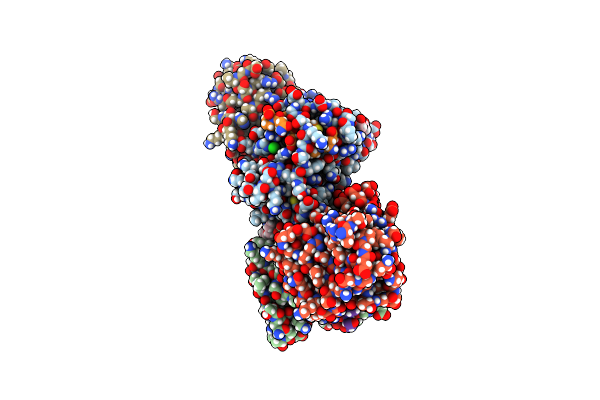 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-12-25 Classification: PROTEIN BINDING Ligands: ACT, CL, 1PE |
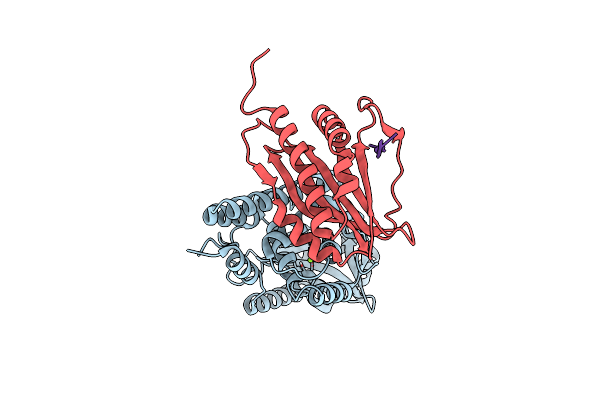 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2019-12-25 Classification: SIGNALING PROTEIN Ligands: MG |
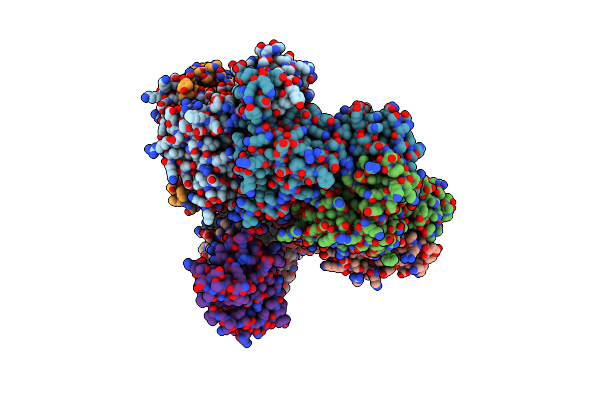 |
Organism: Escherichia coli ms 115-1, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2019-12-25 Classification: PROTEIN BINDING Ligands: ATP, MG |
 |
Organism: Marine metagenome
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-12-25 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-12-25 Classification: DNA BINDING PROTEIN/RNA |
 |
Organism: Escherichia coli ms 115-1, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2019-12-25 Classification: SIGNALING PROTEIN Ligands: MG, CL, ACT, AMP |
 |
Organism: Vibrio metoecus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-12-25 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio metoecus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-12-25 Classification: DNA BINDING PROTEIN Ligands: SO4 |

