Search Count: 13
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm557
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: RL7, FMN, ORO |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm483
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: RLA, FMN, ORO |
 |
Plasmodium Falciparum Dihydroorotate Dehydrogenase Dhodh In Complex With 3,6-Dimethyl-N-(4-(Trifluoromethyl)Phenyl)-(1,2)Oxazolo(5,4-D)Pyrimidin-4-Amine
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FMN, ORO, F1T |
 |
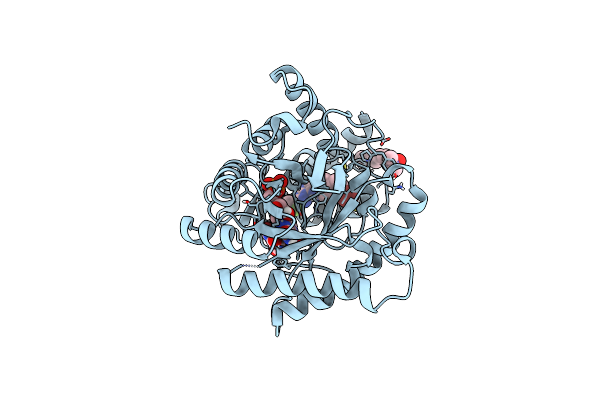
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm421
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-09-28 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: 78Z, FMN, ORO, LDA, GOL |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bounded With Dsm422 (Tetrahydro-2-Naphthyl And 2-Indanyl Triazolopyrimidine)
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2016-05-18 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: 5Y5, FMN, ORO, LDA |
 |
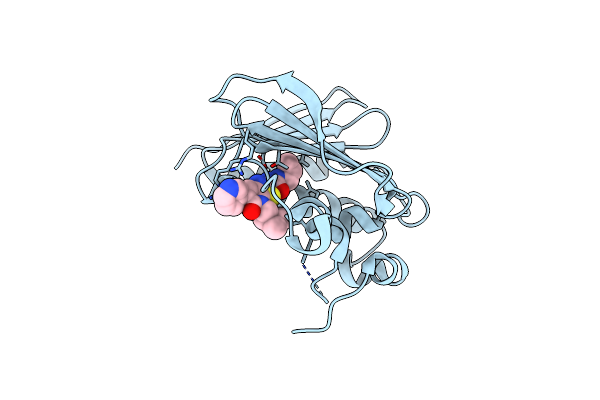
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2014-03-05 Classification: HYDROLASE Ligands: ZN, CA, NA, AZ4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-03-05 Classification: HYDROLASE Ligands: ZN, CA, NA, AZ6 |
 |
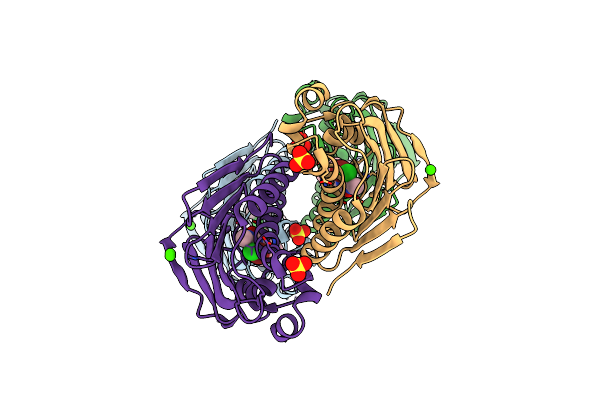
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-11-20 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 2B7 |
 |
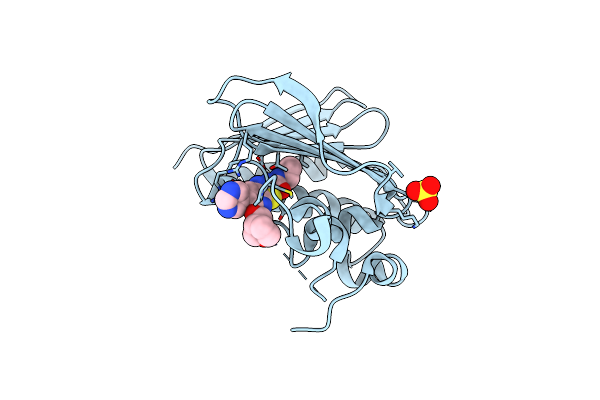
Optimisation Of Pyrroleamides As Mycobacterial Gyrb Atpase Inhibitors: Structure Activity Relationship And In Vivo Efficacy In The Mouse Model Of Tuberculosis
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-10-30 Classification: ISOMERASE Ligands: CA, RWX, SO4 |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-10-16 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: SO4, 28F |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-10-16 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 28G |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-05-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, NFH, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-05-16 Classification: HYDROLASE Ligands: CA, STN, BUM, ZN |

