Search Count: 18
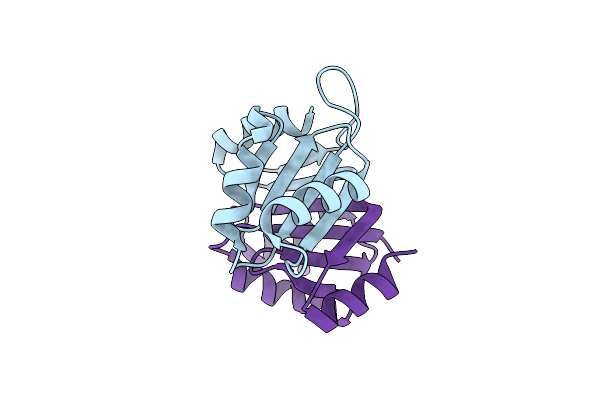 |
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-08-16 Classification: HYDROLASE |
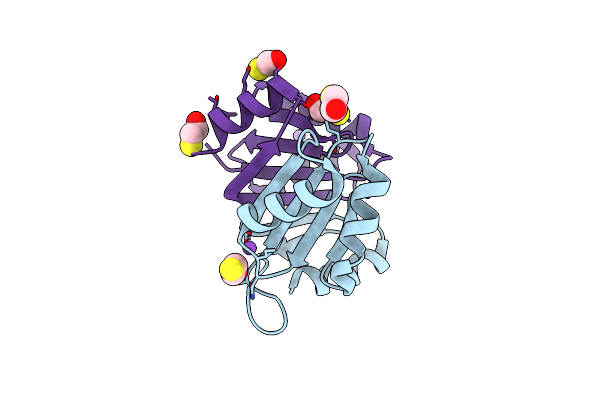 |
Crystal Structure Of A Selenomethionine-Labeled Bpsl1038 From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: BME, NA |
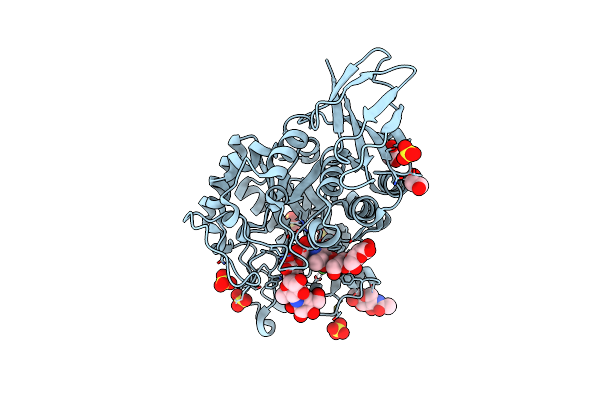 |
Structural And Functional Characterisation Of Three Novel Fungal Amylases With Enhanced Stability And Ph Tolerance
Organism: Thamnidium elegans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: NAG, SO4, EDO, CA |
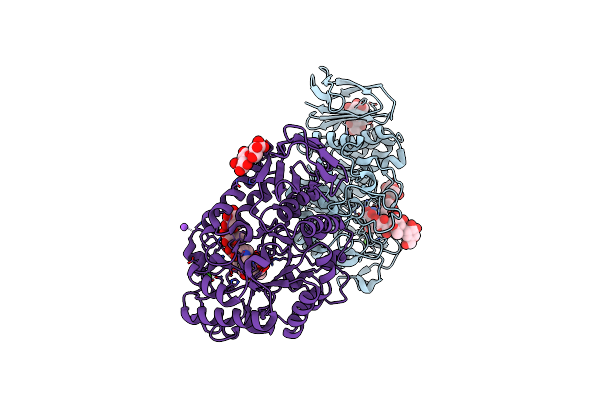 |
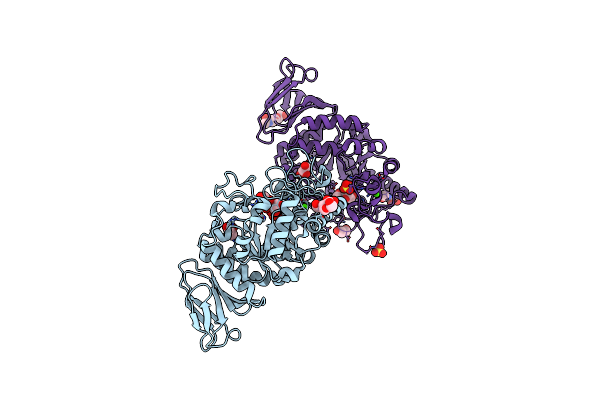
Structural And Functional Characterisation Of Three Novel Fungal Amylases With Enhanced Stability And Ph Tolerance.
Organism: Cordyceps farinosa
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: CA, NA |
 |
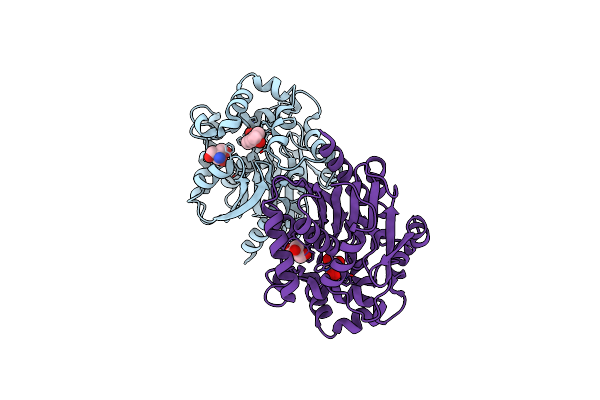
Structural And Functional Characterisation Of Three Novel Fungal Amylases With Enhanced Stability And Ph Tolerance
Organism: Rhizomucor pusillus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: CA, NAG, GOL, SO4 |
 |
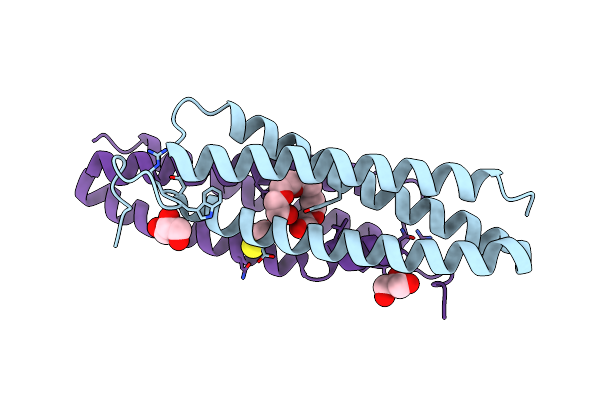
Crystal Structure Of The Common Ancestor Of Haloalkane Dehalogenases And Renilla Luciferase (Anchld-Rluc)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: OXY, MPD, TRS |
 |
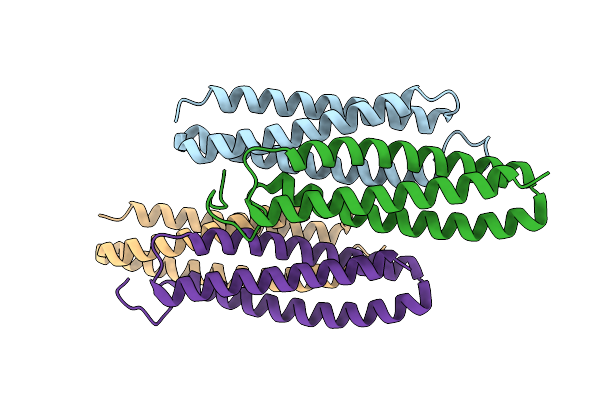
Organism: Dermatophagoides farinae
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2019-03-13 Classification: ALLERGEN Ligands: 1PE, GOL, BME |
 |
Organism: Dermatophagoides farinae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-03-13 Classification: ALLERGEN |
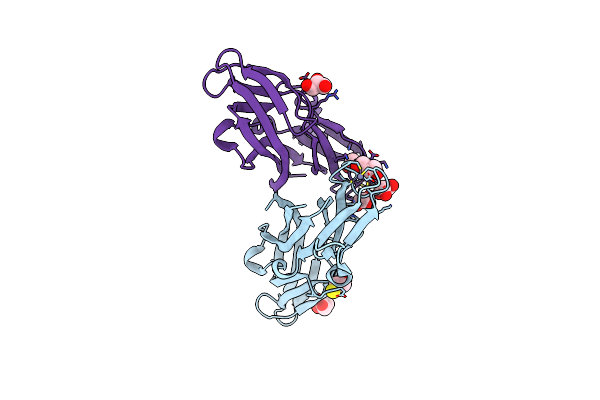 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-01 Classification: UNKNOWN FUNCTION Ligands: GOL, CL, ZN, BME |
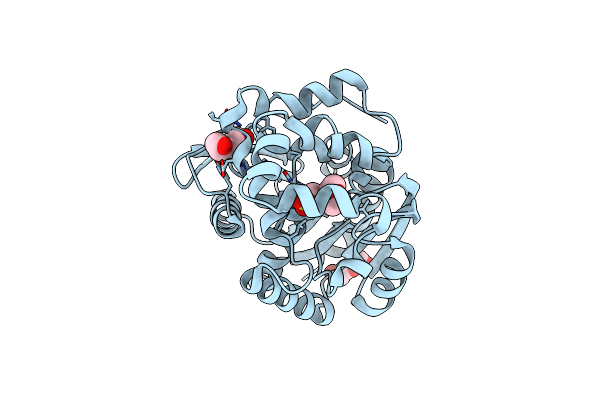 |
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2017-03-29 Classification: HYDROLASE Ligands: MES, PEG |
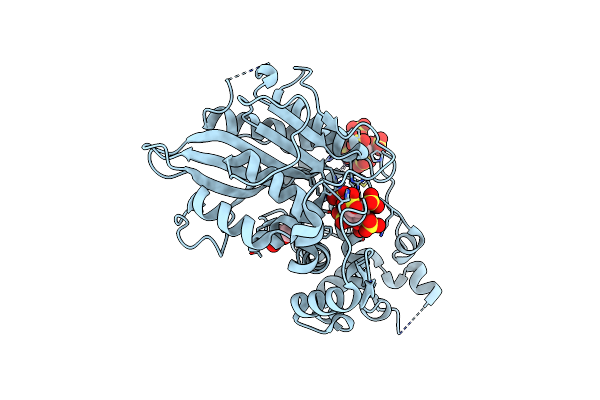 |
Organism: Hafnia alvei
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: IHS, PEG, K |
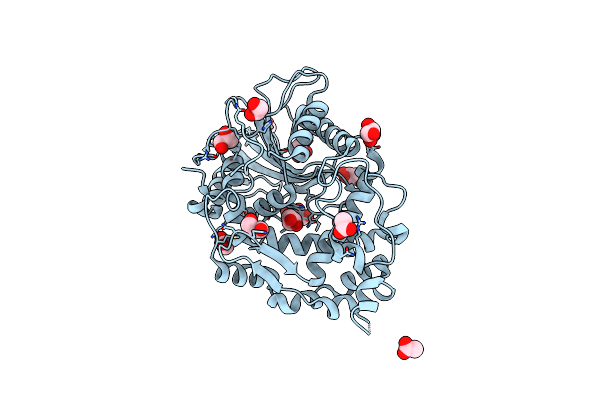 |
Organism: Hafnia alvei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: GOL, ACT |
 |
Organism: Hafnia alvei
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: TLA, NA, K, CL |
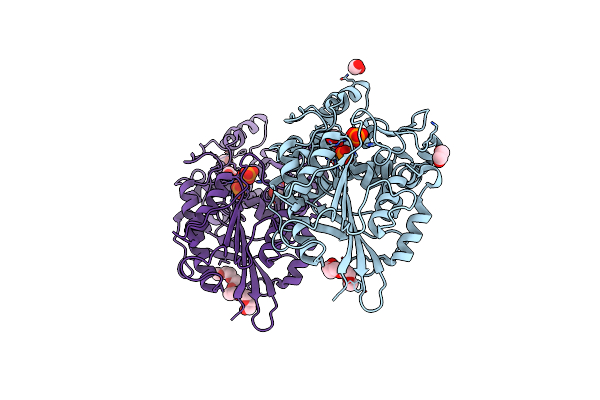 |
Organism: Yersinia kristensenii
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: PO4, EDO, TOE, P15 |
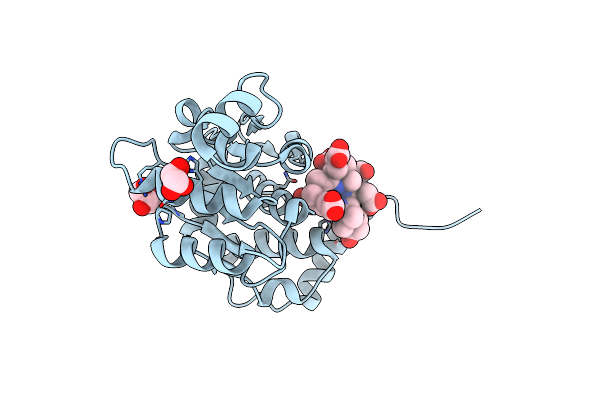 |
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2012-10-17 Classification: ISOMERASE Ligands: 0UK, GOL |
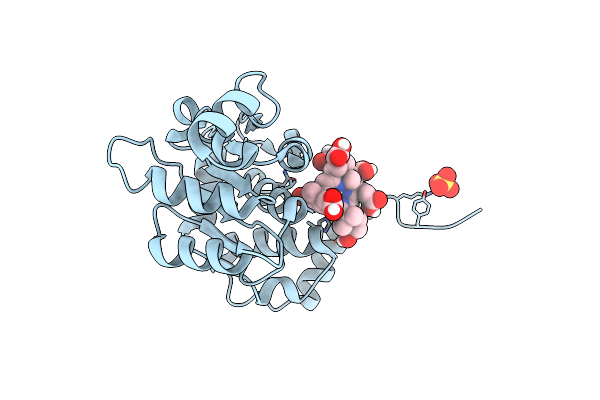 |
Crystal Structure Of Cobh (Precorrin-8X Methyl Mutase) Complexed With C5 Desmethyl-Hba
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-09-19 Classification: ISOMERASE Ligands: P8X, SO4 |
 |
Crystal Structure Of C-Terminal Domain Of Precorrin-6Y C5,15-Methyltransferase From Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-06-08 Classification: TRANSFERASE Ligands: SAH, GOL, PG4 |
 |
Solution Structure Determination Of A P53 Mutant Dimerization Domain, Nmr, Minimized Average Structure
|

