Search Count: 13
 |
Crystal Structure Of Proteinase K Nanocrystals By Electron Diffraction With A 20 Micrometre C2 Condenser Aperture
Organism: Parengyodontium album
Method: ELECTRON CRYSTALLOGRAPHY Resolution:2.70 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Proteinase K Lamella By Electron Diffraction With A 50 Micrometre C2 Condenser Aperture
Organism: Parengyodontium album
Method: ELECTRON CRYSTALLOGRAPHY Resolution:2.00 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Proteinase K Lamellae By Electron Diffraction With A 20 Micrometre C2 Condenser Aperture
Organism: Parengyodontium album
Method: ELECTRON CRYSTALLOGRAPHY Resolution:2.40 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CA |
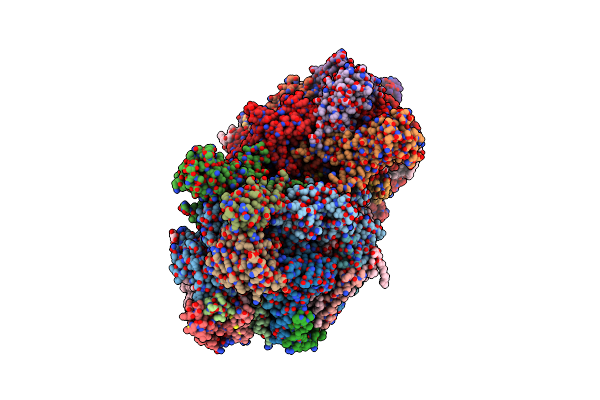 |
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-11-23 Classification: ELECTRON TRANSPORT Ligands: OEX, FE2, SQD, CL, CLA, PHO, BCR, PL9, LMG, UNL, LHG, BCT, DGD, HEM, HEC |
 |
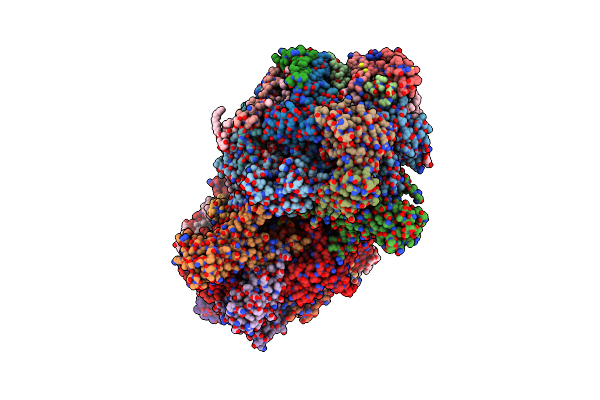
Nh3-Bound Rt Xfel Structure Of Photosystem Ii 500 Ms After The 2Nd Illumination (2F) At 2.8 A Resolution
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-23 Classification: ELECTRON TRANSPORT Ligands: OEX, FE2, LMG, CL, CLA, PHO, BCR, PL9, SQD, UNL, BCT, LHG, DGD, HEM, HEC |
 |
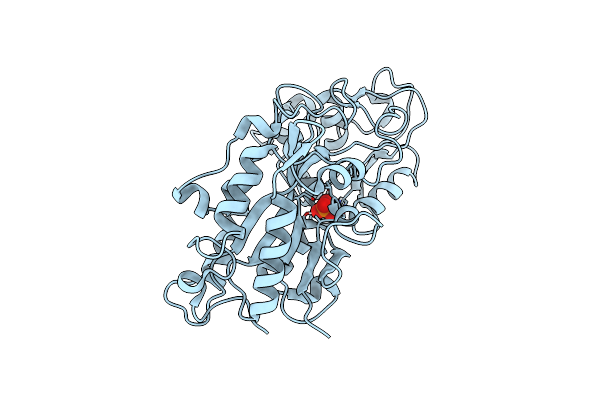
Room Temperature Xfel Structure Of The Native, Doubly-Illuminated Photosystem Ii Complex
Organism: Thermosynechococcus elongatus (strain bp-1), Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-11-23 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, CL, BCT, CLA, PHO, BCR, PL9, SQD, LHG, UNL, LMG, DGD, HEM, HEC |
 |
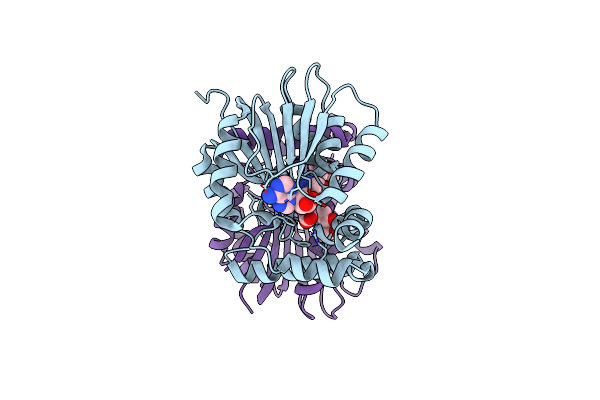
Organism: Stenotrophomonas maltophilia r551-3
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2016-05-04 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Crystal Structure Of A Putative Methyltransferase Cmoa In Complex With A Novel Sam Derivative
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2013-05-29 Classification: TRANSFERASE Ligands: GEK, MPD |
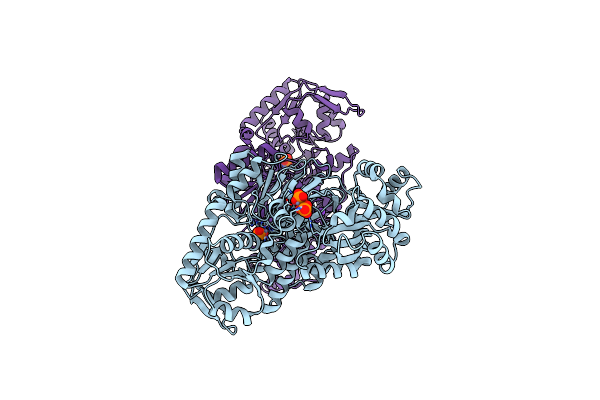 |
Structure Of The Archaeal Beta-Casp Protein With N-Terminal Kh Domains From Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2011-05-25 Classification: HYDROLASE Ligands: ZN, PO4, K |
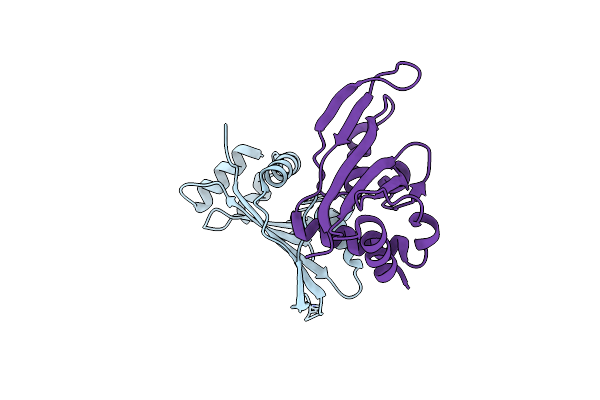 |
Structure Of Mth689, A Duf54 Protein From Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-09-01 Classification: UNKNOWN FUNCTION Ligands: NI, CL |
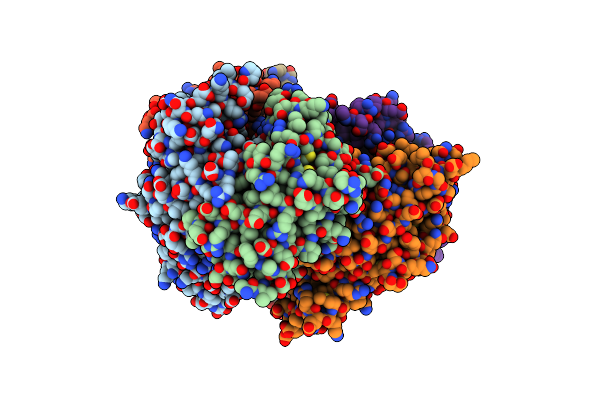 |
The Structure Of Methanothermobacter Thermautotrophicus Exosome Core Assembly
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2010-04-28 Classification: HYDROLASE Ligands: PO4 |
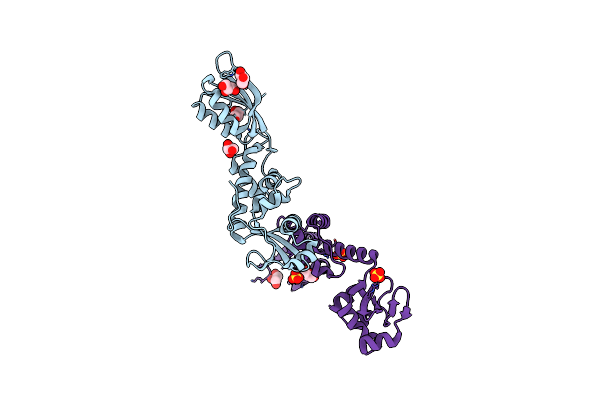 |
Crystal Structure Of Mthsbds, The Homologue Of The Shwachman-Bodian- Diamond Syndrome Protein In The Euriarchaeon Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-06-02 Classification: RNA BINDING PROTEIN Ligands: GOL, SO4, CL |
 |
Crystal Structure Of Bacillus Anthracis Thii, A Trna-Modifying Enzyme Containing The Predicted Rna-Binding Thump Domain
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-11-30 Classification: RNA BINDING PROTEIN Ligands: AMP |

