Search Count: 248
 |
Organism: Agrobacterium tumefaciens (strain c58)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: SAH, SO4 |
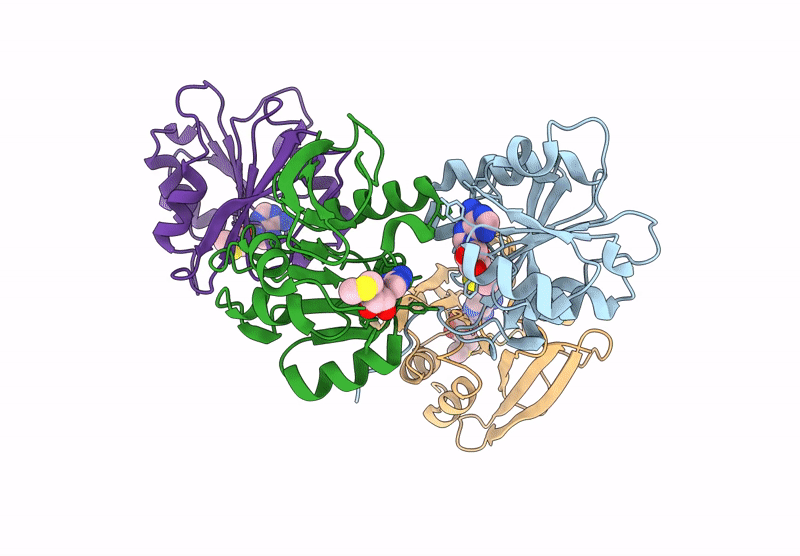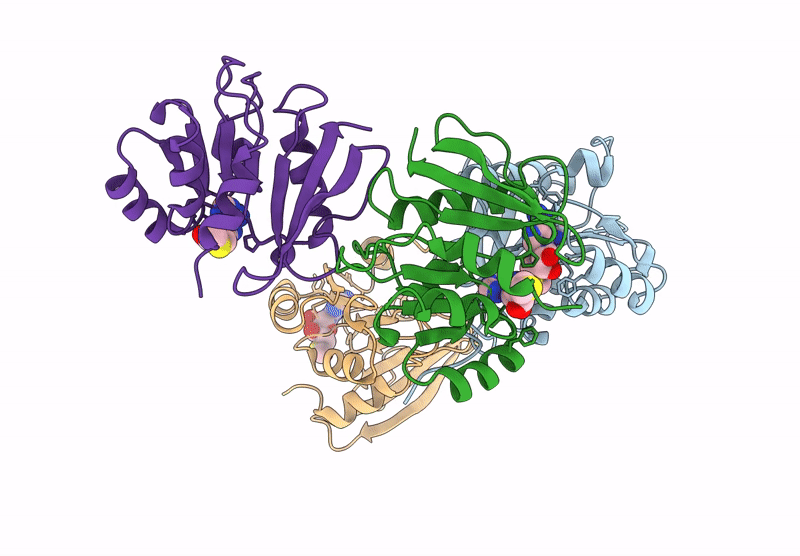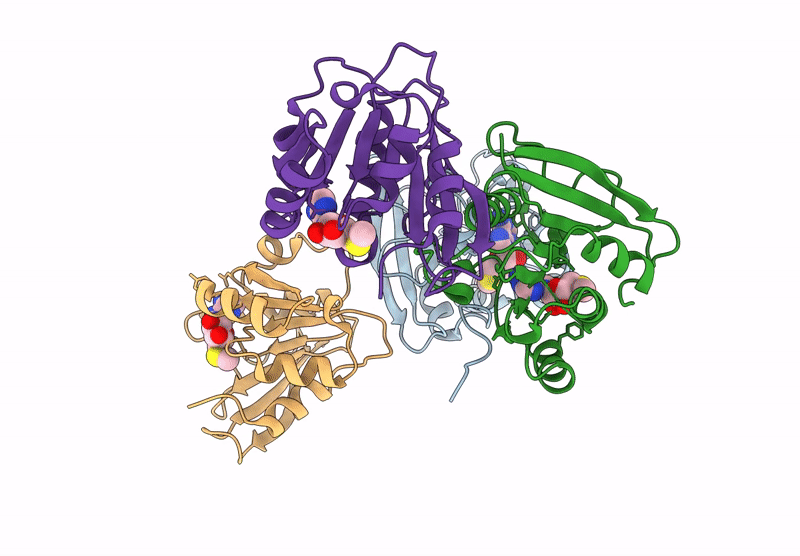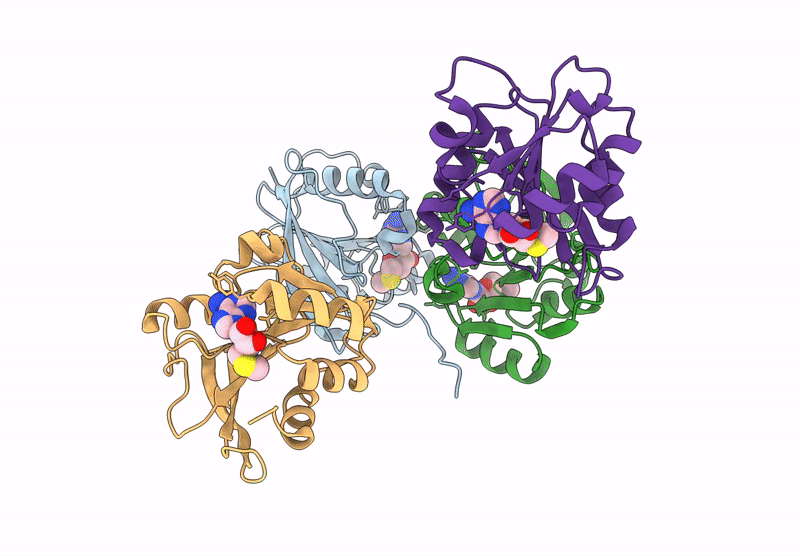 |
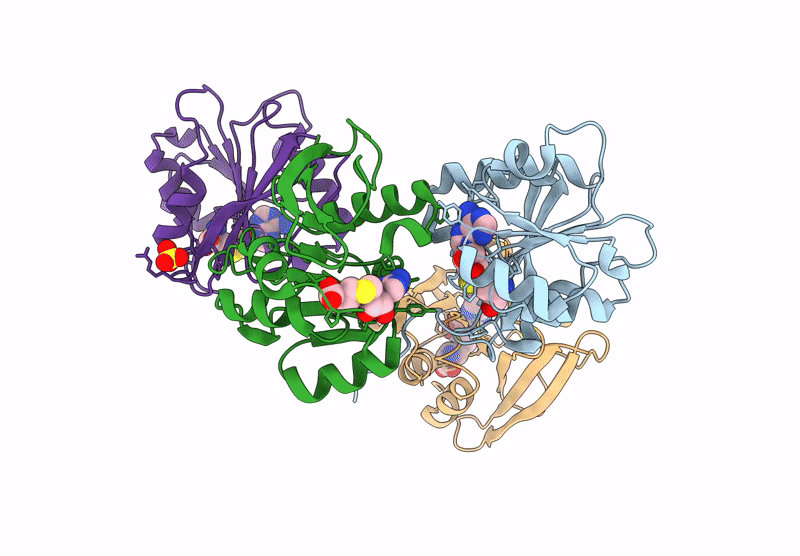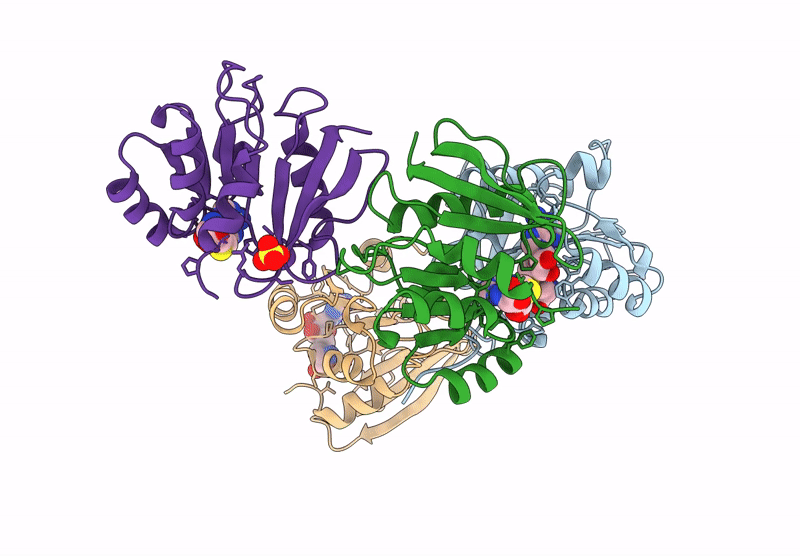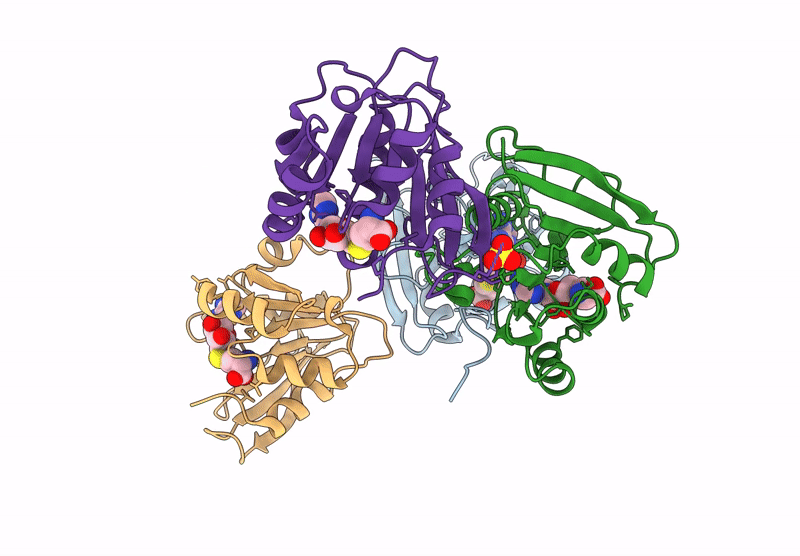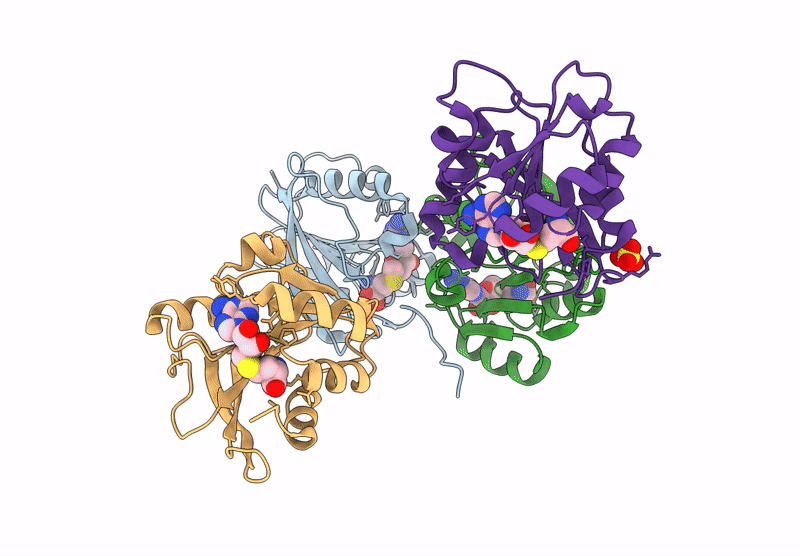
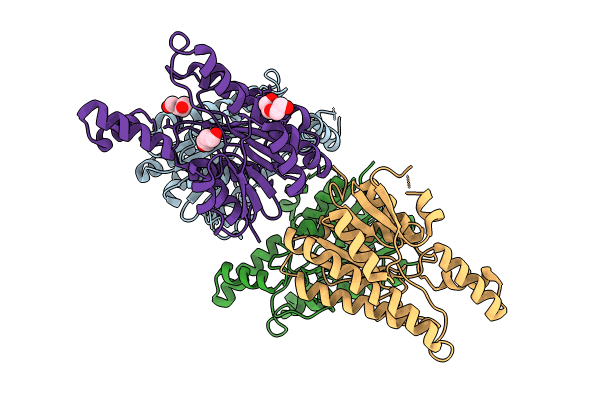
Crystal Structure Of Agrobacterium Tumefaciens Pmta With 5'-Methylthioadenosine
Organism: Agrobacterium tumefaciens (strain c58)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: MTA |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase From Herbaspillum Huttiense (Apo Form)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: PEG |
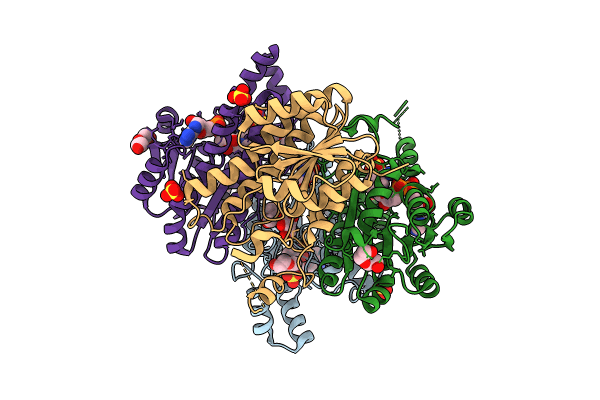 |
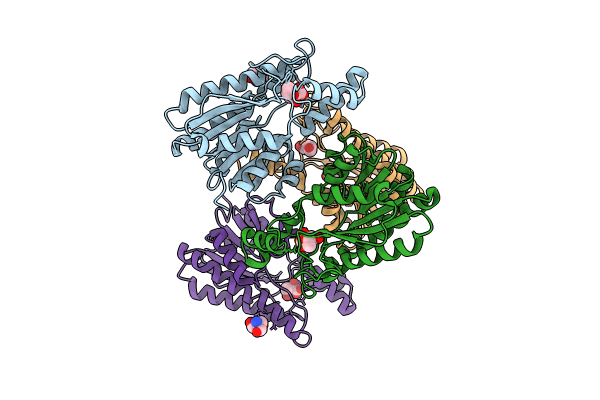
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To Nad(H) And Sulfate Ion
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, PEG, TRS, GOL |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To L-Kdf Or L-2,4-Dkdf
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: TRS, A1LXA, A1LXB, PEG |
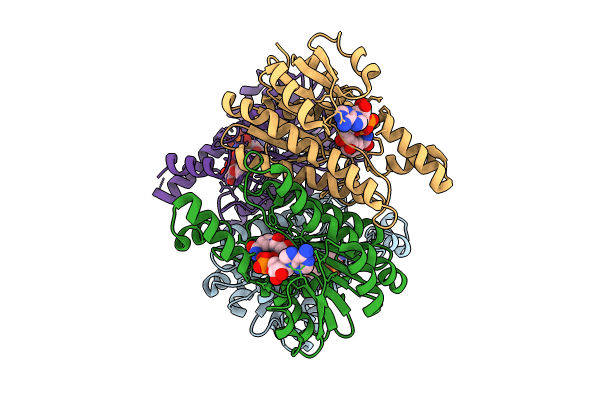 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To L-2,4-Dkdf And Nadh
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, A1LXB |
 |
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: TRS, A1LXD, PEG |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-02-14 Classification: LYASE Ligands: TVJ, GOL, EDO |
 |
Discovery And Characterization Of A New Carbonyl Reductase From Rhodotorula Toluroides Reducing Fluoroketones, And X-Ray Analysis Of The Variant By Rational Engineering
Organism: Rhodotorula toruloides
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: MG |
 |
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: HBX, CL, SO4, GOL |
 |
Hydroxynitrile Lyase From The Millipede, Oxidus Gracilis Bound With (R)-(+)-Alpha-Hydroxybenzene-Acetonitrile
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: MXN, SO4 |
 |
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: CNH, CL, SO4 |
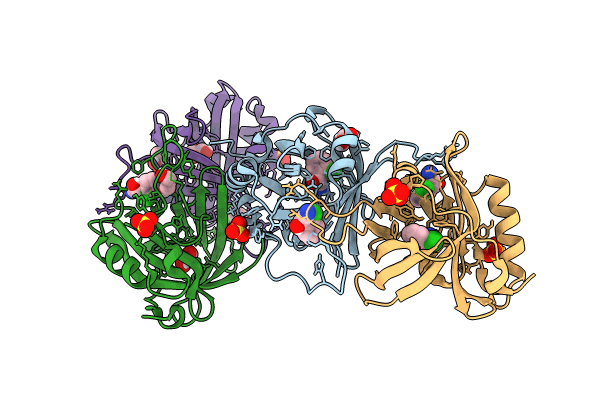 |
Hydroxynitrile Lyase From The Millipede, Oxidus Gracilis Complexed With (R)-2-Chloromandelonitrile
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: IJ5, GOL, SO4 |
 |
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-17 Classification: LYASE Ligands: SO4, CL |
 |
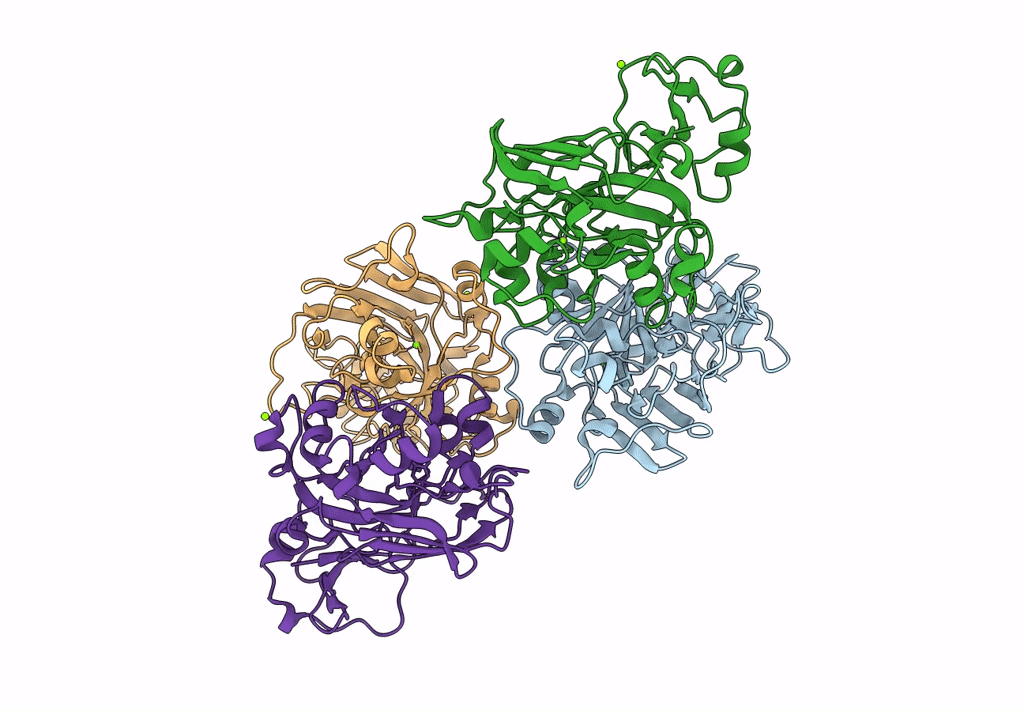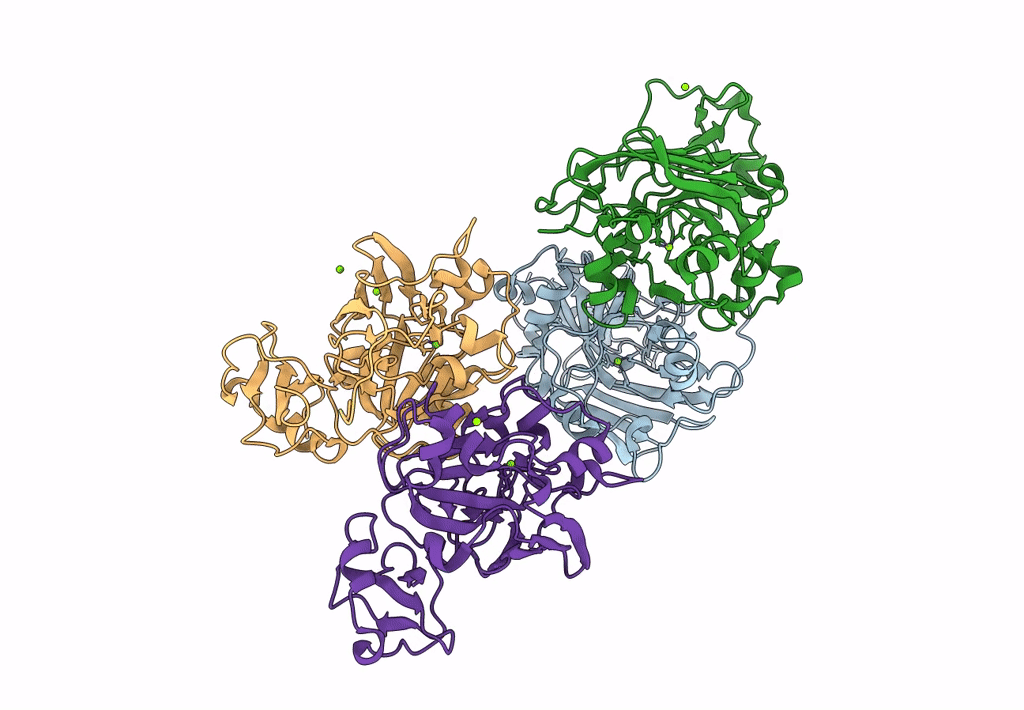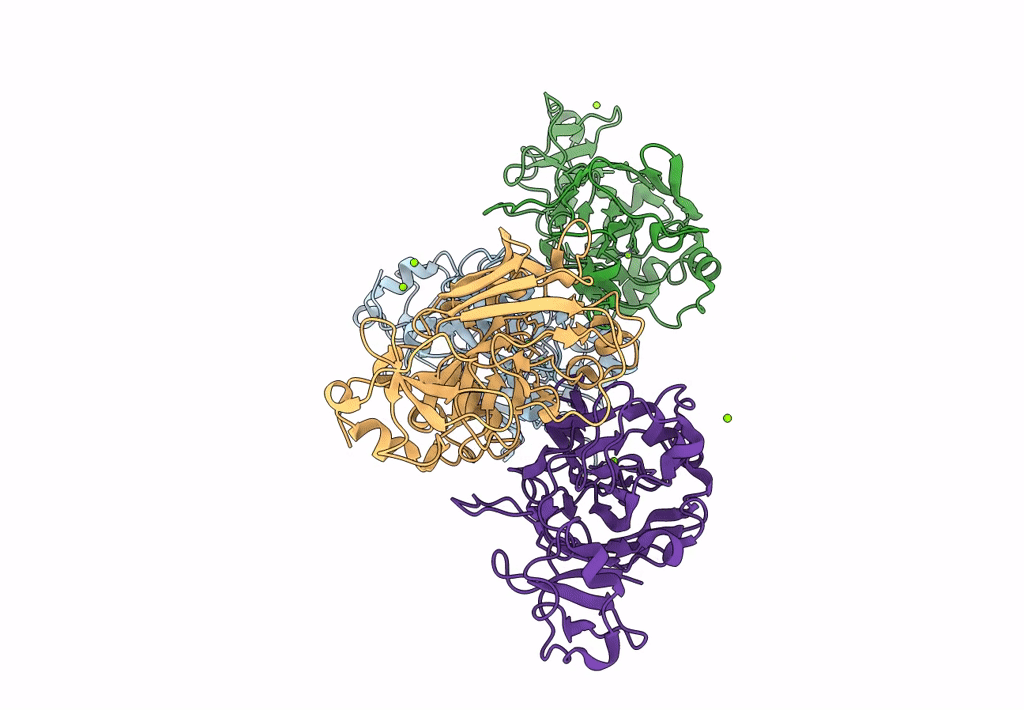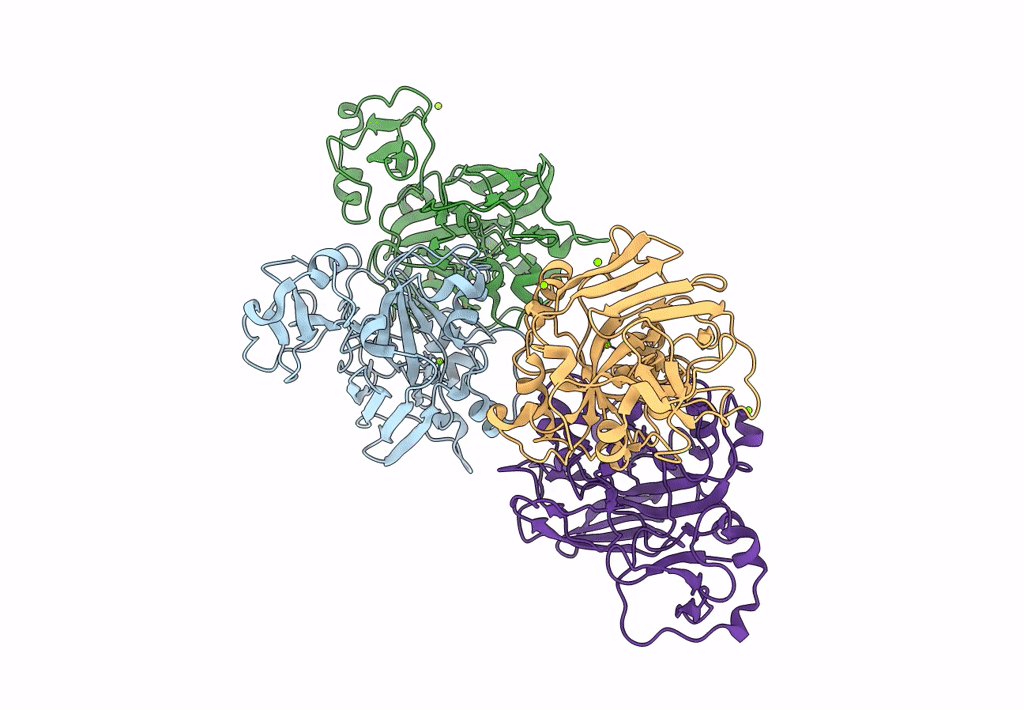
Xylitol Dehydrogenase S96C/S99C/Y102C Mutant(Thermostabilized Form) From Pichia Stipitis
Organism: Scheffersomyces stipitis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: ZN, GOL, SO4, PEG |
 |
Crystal Structure Of L-2,4-Diketo-3-Deoxyrhamnonate Hydrolase From Sphingomonas Sp. (Apo-Form)
Organism: Sphingomonas sp. ska58
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-02-08 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of L-2,4-Diketo-3-Deoxyrhamnonate Hydrolase From Sphingomonas Sp. (Pyruvate Bound-Form)
Organism: Sphingomonas sp. ska58
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-02-08 Classification: HYDROLASE Ligands: MG, PYR |
 |
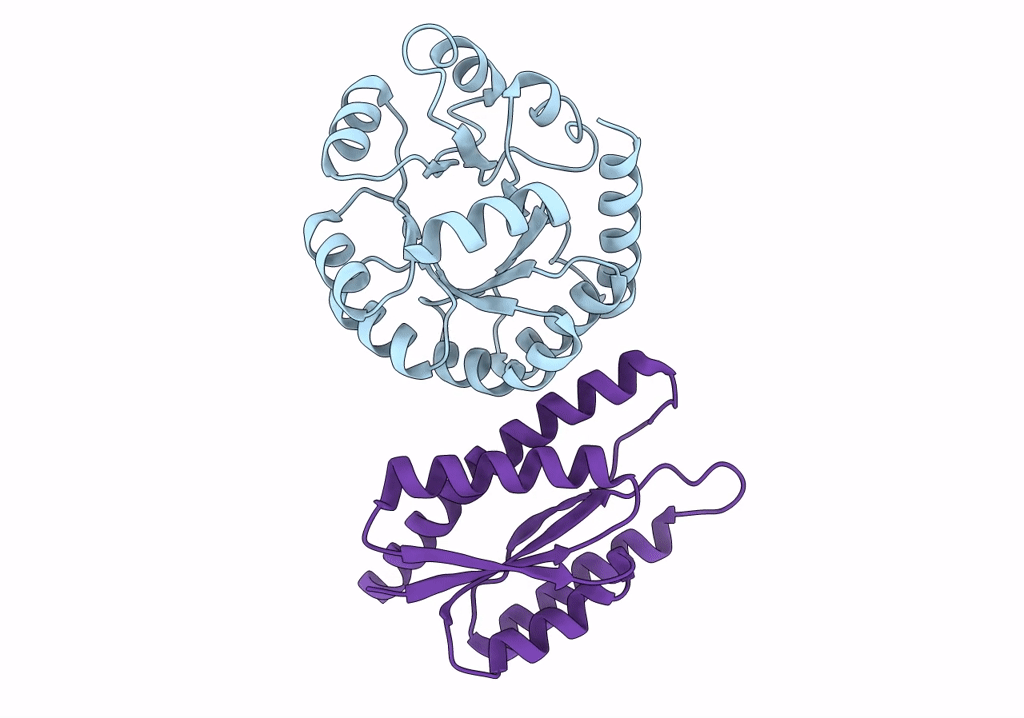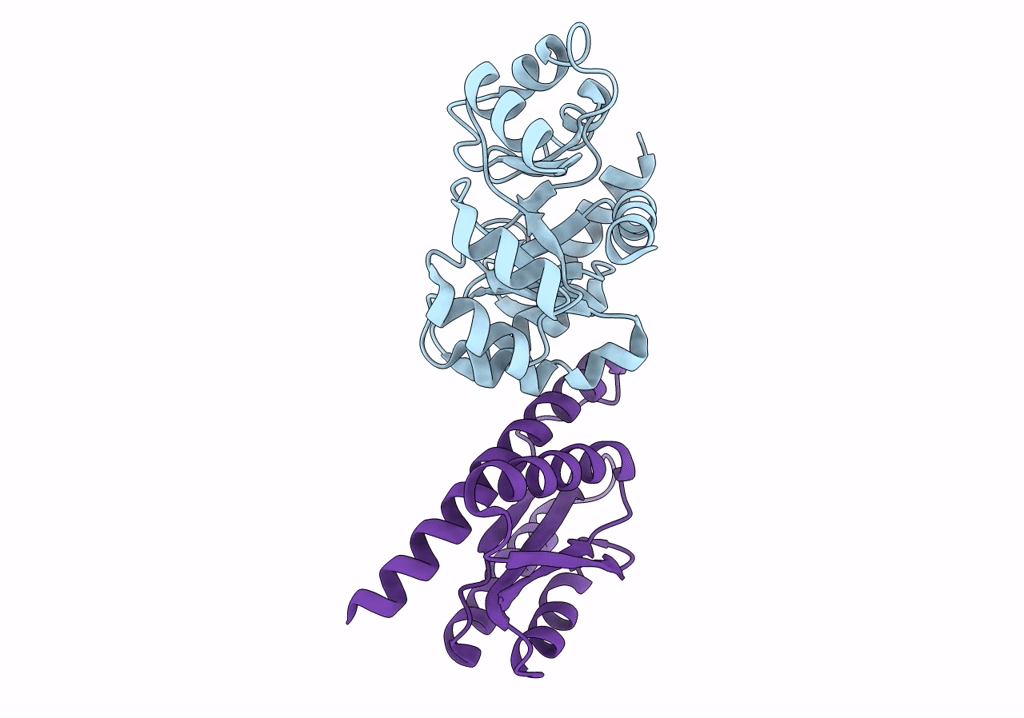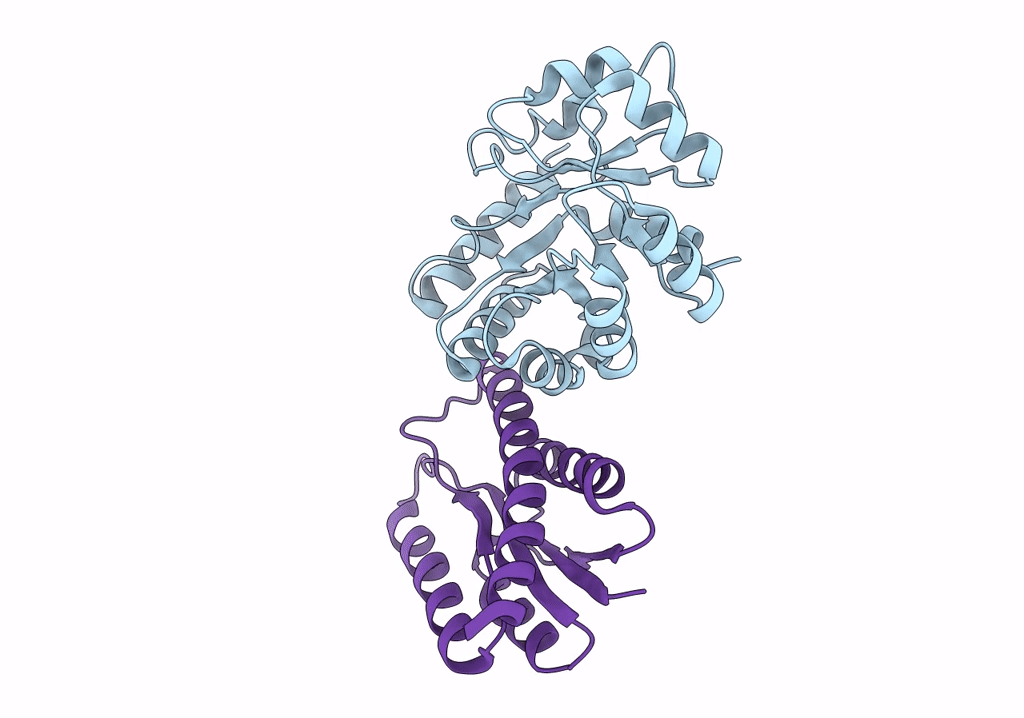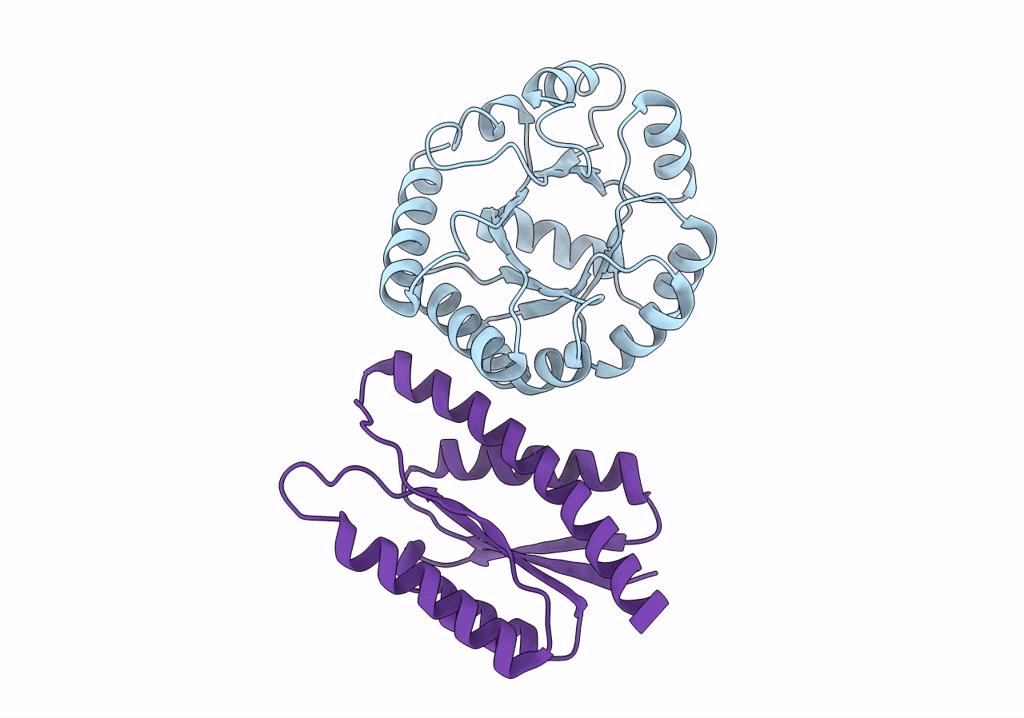
Lassa Virus Glycoprotein Construct(Josiah Gpcysr4) Recovered From Gpc-I53-50 Nanoparticle By Localized Reconstruction
Organism: Lassa mammarenavirus
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: VIRAL PROTEIN |
 |
Lassa Virus Glycoprotein Construct (Josiah Gpc-I53-50A) In Complex With Lava01 Antibody
Organism: Lassa mammarenavirus, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |

