Search Count: 335
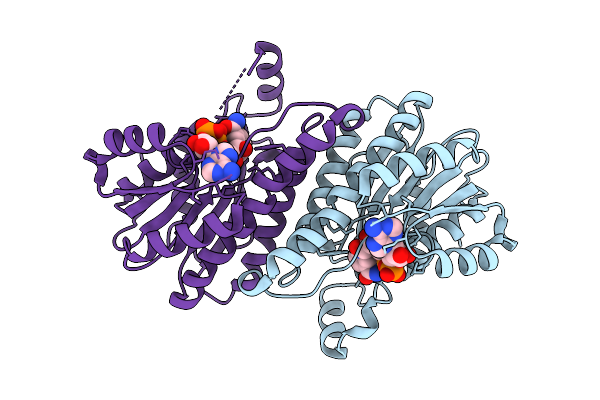 |
Crystal Structure Of L-2-Keto-3-Deoxypentonate 4-Dehydrogenase Bound To Nad(H)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crysral Structure Of 2-Keto-3-Deoxypentonate 4-Dehydrogenase From Herbaspirillum Huttiense (Apo Form)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE |
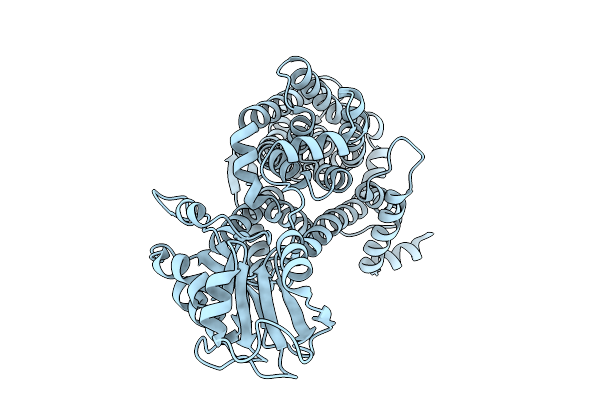 |
X-Ray Crystal Structure Of Asp/Ala Exchanger Aspt At Outward-Facing Conformation
Organism: Tetragenococcus halophilus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Tetragenococcus halophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: TRANSPORT PROTEIN Ligands: ASP |
 |
Organism: Tetragenococcus halophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: TRANSPORT PROTEIN |
 |
Organism: Tetragenococcus halophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: TRANSPORT PROTEIN Ligands: ASP |
 |
Organism: Tetragenococcus halophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
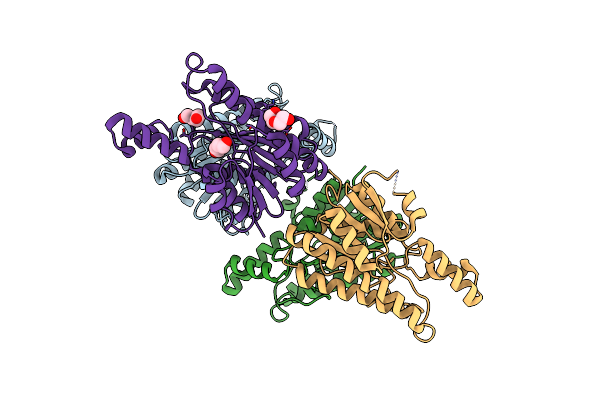 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase From Herbaspillum Huttiense (Apo Form)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: PEG |
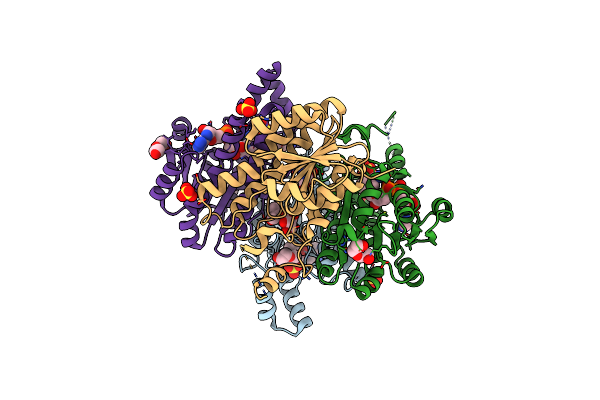 |
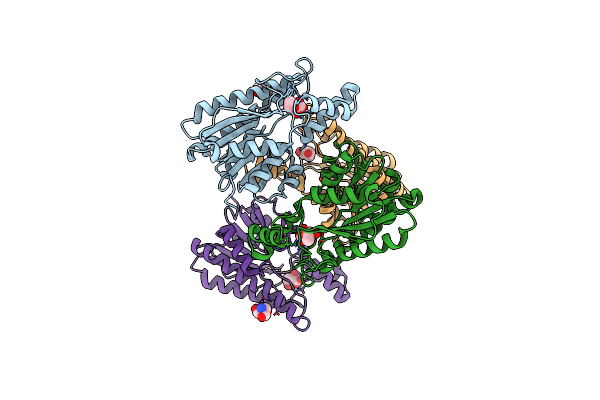
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To Nad(H) And Sulfate Ion
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, PEG, TRS, GOL |
 |
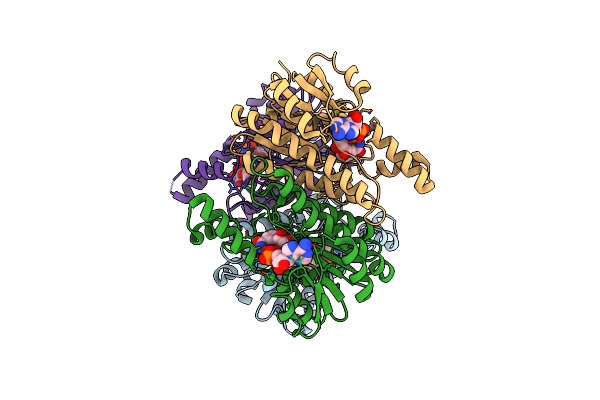
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To L-Kdf Or L-2,4-Dkdf
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: TRS, A1LXA, A1LXB, PEG |
 |
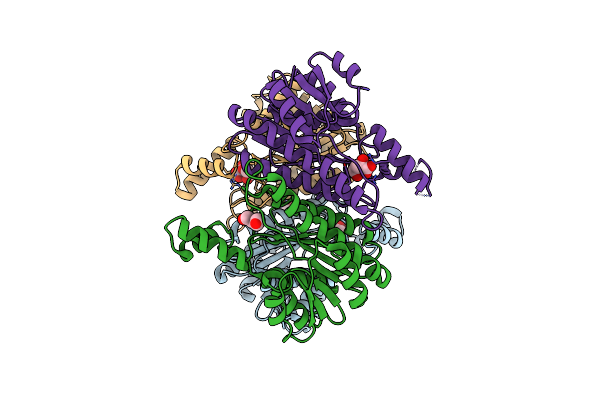
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To L-2,4-Dkdf And Nadh
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, A1LXB |
 |
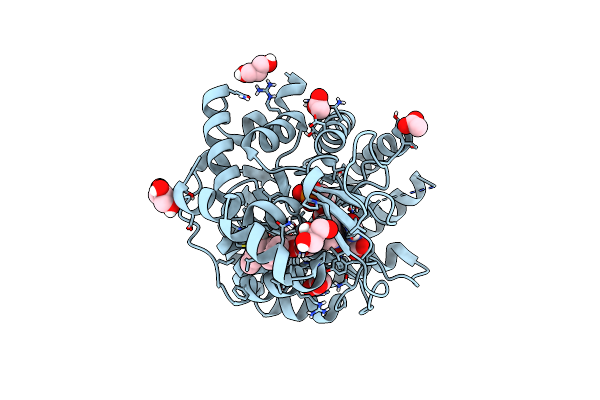
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To D-Kdp
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: TRS, A1LXD, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-05-22 Classification: OXIDOREDUCTASE Ligands: ORO, FMN, GOL, ACT, SO4, A1LYU |
 |
Cryo-Em Structure Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Form A)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structures Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Form B)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structures Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Substate A)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structure Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Substate B)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structure Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Substate C)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structure Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Substate D)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |

