Search Count: 155
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With 4456
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1BYI |
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With Sj6675
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1BYV |
 |
Cryo-Em Structure Of Cryptococcus Neoformans Trehalose-6-Phosphate Synthase Homotetramer In Apo Form
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Cryptococcus Neoformans Trehalose-6-Phosphate Synthase Homotetramer In Complex With Uridine Diphosphate And Glucose-6-Phosphate
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: TRANSFERASE Ligands: UDP, G6P |
 |
Structure Of Trehalose-6-Phosphate Phosphatase From Cryptococcus Neoformans
Organism: Cryptococcus neoformans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-06-15 Classification: HYDROLASE Ligands: MG, BME |
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Phosphatase N-Terminal Domain
Organism: Candida albicans (strain sc5314 / atcc mya-2876)
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2016-06-15 Classification: HYDROLASE |
 |
Structure Of C. Albicans Trehalose-6-Phosphate Phosphatase C-Terminal Domain
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-06-15 Classification: HYDROLASE Ligands: MG, BEF |
 |
Structure Of Aspergillus Fumigatus Trehalose-6-Phosphate Phosphatase Crystal Form 1
Organism: Neosartorya fumigata
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2016-06-15 Classification: HYDROLASE |
 |
Structure Of Aspergillus Fumigatus Trehalose-6-Phosphate Phosphatase Crystal Form 2
Organism: Neosartorya fumigata
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-06-15 Classification: HYDROLASE Ligands: MG |
 |
Structure Of Aspergillus Fumigatus Trehalose-6-Phosphate Phosphatase Crystal Form 3
Organism: Neosartorya fumigata
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-06-15 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of A Terpene Synthase From Streptomyces Lydicus, Target Efi-540129
Organism: Streptomyces lydicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-08 Classification: TRANSFERASE |
 |
Crystal Structure Of A Isoprenoid Synthase Family Member From Thermotoga Neapolitana Dsm 4359, Target Efi-509458
Organism: Thermotoga neapolitana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-11-19 Classification: TRANSFERASE |
 |
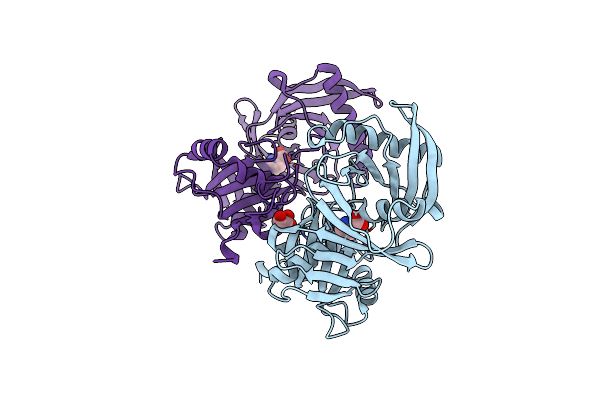
Crystal Structure Of Sugar Transporter Atu4361 From Agrobacterium Fabrum C58, Target Efi-510558, With Bound Maltotriose
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2014-07-16 Classification: TRANSPORT PROTEIN |
 |
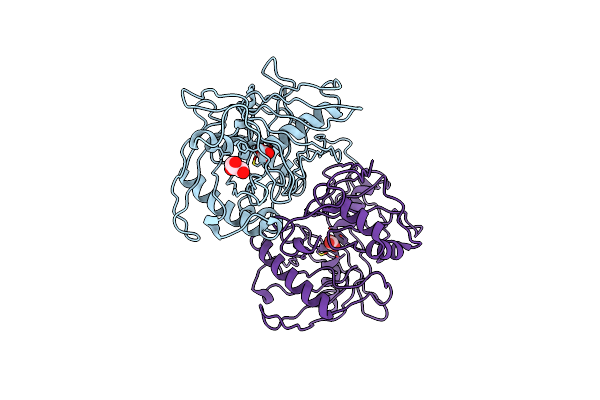
Crystal Structure Of A 4-Hydroxyproline Epimerase From Burkholderia Multivorans Atcc 17616, Target Efi-506586, Open Form, With Bound Pyrrole-2-Carboxylate
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-07 Classification: ISOMERASE Ligands: GOL, PYC |
 |
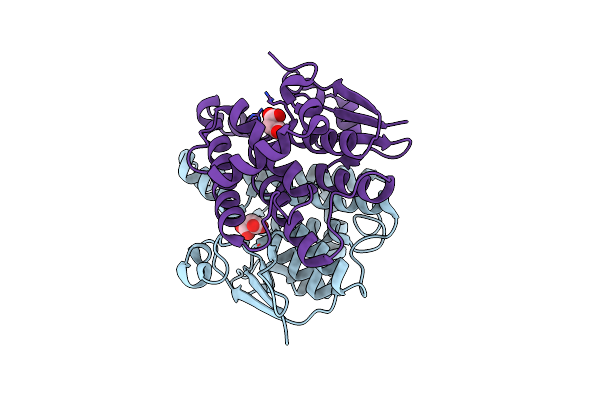
Crystal Structure Of Probable Proline Racemase From Agrobacterium Radiobacter K84, Target Efi-506561, With Bound Carbonate
Organism: Agrobacterium radiobacter k84
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-30 Classification: ISOMERASE Ligands: BCT, GOL |
 |
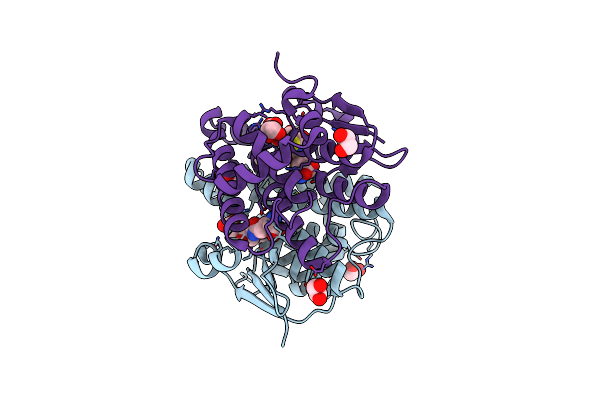
Crystal Structure Of Maleylacetoacetate Isomerase From Methylobacteriu Extorquens Am1 With Bound Malonate (Target Efi-507068)
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-04-09 Classification: TRANSFERASE Ligands: MLA, CL |
 |
Crystal Structure Of Maleylacetoacetate Isomerase From Methylobacteriu Extorquens Am1 With Bound Malonate And Gsh (Target Efi-507068)
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-09 Classification: TRANSFERASE Ligands: MLA, GSH, EDO |
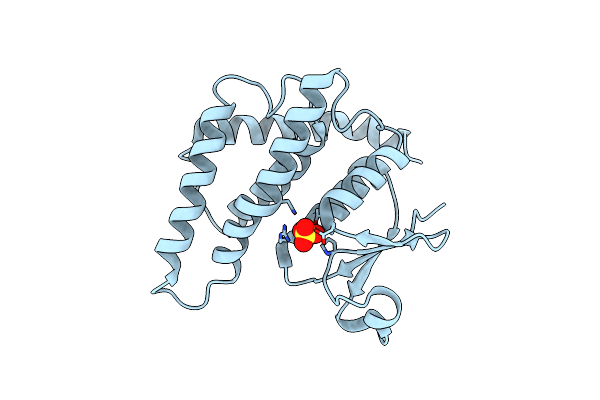 |
Crystal Structure Of A Glutathione S-Transferase From Pichia Kudriavzevii (Issatchenkia Orientalis), Target Efi-501747
Organism: Pichia kudriavzevii
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2014-01-15 Classification: TRANSFERASE Ligands: SO4 |
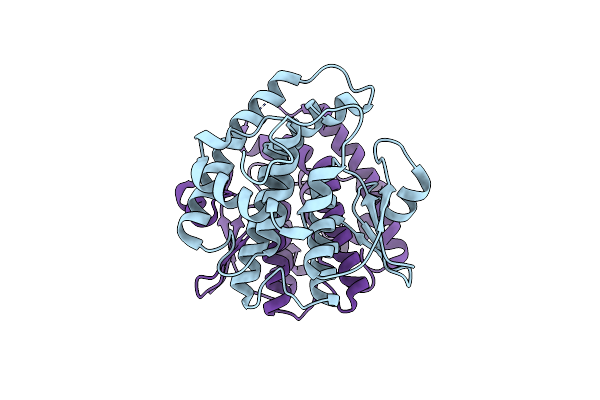 |
Crystal Structure Of A Glutathione S-Transferase From Rhodospirillum Rubrum F11, Target Efi-507460
Organism: Rhodospirillum rubrum f11
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-01-08 Classification: TRANSFERASE |
 |
Crystal Structure Of Glutathione Transferase Smc00097 From Sinorhizobium Meliloti, Target Efi-507275, With One Glutathione Bound Per One Protein Subunit
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-11-20 Classification: TRANSFERASE Ligands: GSH |

