Search Count: 13
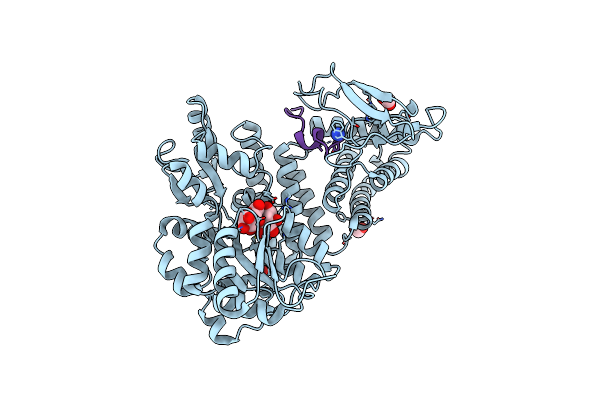 |
Crystal Structure Of The Human Clr:Ramp2 Extracellular Domain Heterodimer With Bound High-Affinity Adrenomedullin S45R/K46L/S48G/Q50W Variant
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2020-08-05 Classification: MEMBRANE PROTEIN Ligands: FMT, NH2 |
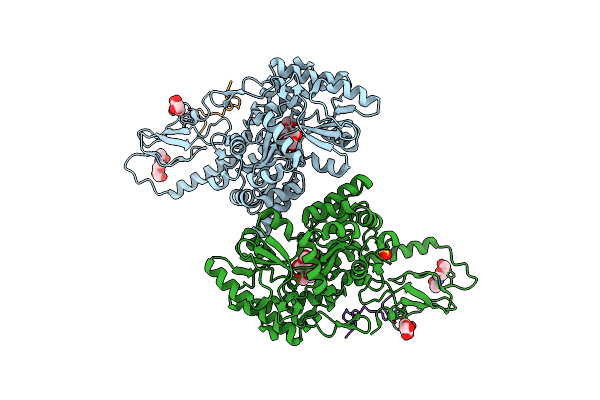 |
Crystal Structure Of N-Glycosylated Human Calcitonin Receptor Extracellular Domain In Complex With Salmon Calcitonin (16-32)
Organism: Escherichia coli, Homo sapiens, Oncorhynchus sp.
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN Ligands: NAG, SO4 |
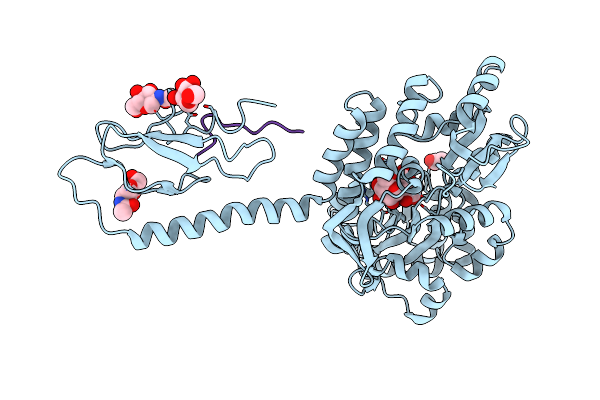 |
Crystal Structure Of N-Glycosylated Human Calcitonin Receptor Extracellular Domain In Complex With Salmon Calcitonin (22-32)
Organism: Escherichia coli, Homo sapiens, Oncorhynchus sp.
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN Ligands: NAG, ACT |
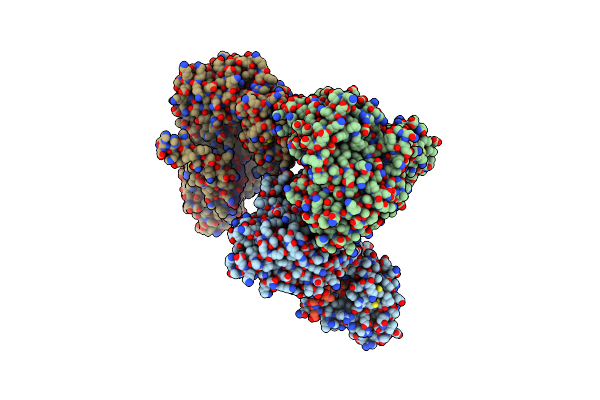 |
Crystal Structure Of The Human Clr:Ramp1 Extracellular Domain Heterodimer In Complex With Adrenomedullin 2/Intermedin
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-09-05 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of The Human Clr:Ramp1 Extracellular Domain Heterodimer With Bound High-Affinity And Altered Selectivity Adrenomedullin Variant
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-01-31 Classification: TRANSPORT PROTEIN / MEMBRANE PROTEIN |
 |
Crystal Structure Of The Clr:Ramp2 Extracellular Domain Heterodimer With Bound Adrenomedullin
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2015-05-20 Classification: MEMBRANE PROTEIN/HORMONE Ligands: EDO |
 |
Crystal Structure Of The Clr:Ramp1 Extracellular Domain Heterodimer With Bound High Affinity Cgrp Analog
Organism: Escherichia coli, Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2015-05-20 Classification: MEMBRANE PROTEIN/HORMONE Ligands: MG |
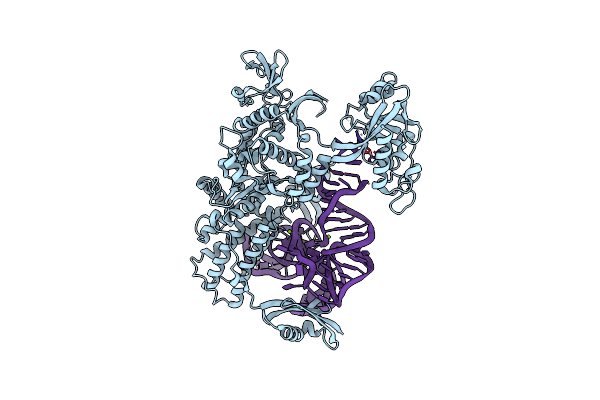 |
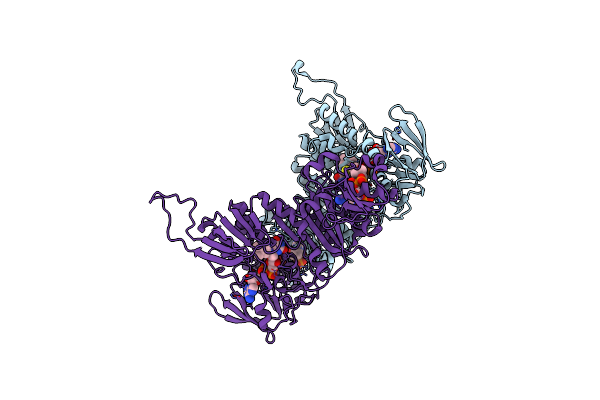
Ternary Complex Of E.Coli Leucyl-Trna Synthetase, Trna(Leu)574 And The Benzoxaborole An3017 In The Editing Conformation
Organism: Escherichia coli, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-17 Classification: LIGASE/RNA Ligands: MG |
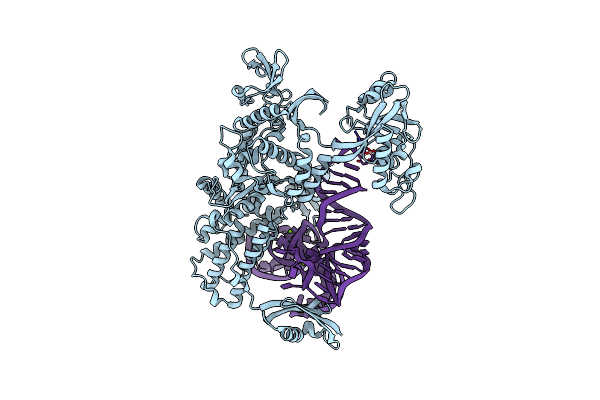 |
Ternary Complex Of E .Coli Leucyl-Trna Synthetase, Trna(Leu) And The Benzoxaborole An3016 In The Editing Conformation
Organism: Escherichia coli, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-04-17 Classification: LIGASE/RNA Ligands: MG |
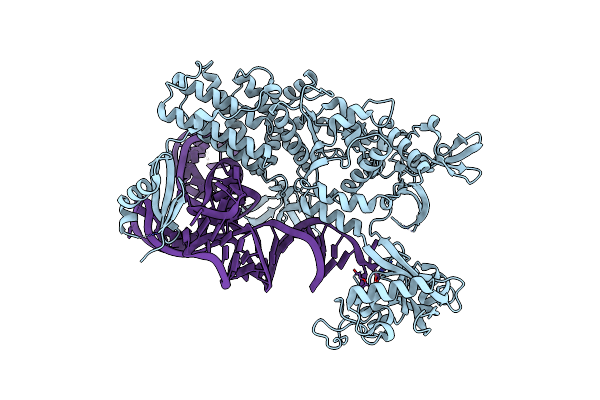 |
Ternary Complex Of E .Coli Leucyl-Trna Synthetase, Trna(Leu) And The Benzoxaborole An3213 In The Editing Conformation
Organism: Escherichia coli, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2013-04-17 Classification: LIGASE/RNA |
 |
Structure Of The Shewanella Loihica Pv-4 Nadh-Dependent Persulfide Reductase C43S/C531S Double Mutant
Organism: Shewanella loihica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: CL, FAD, COA |
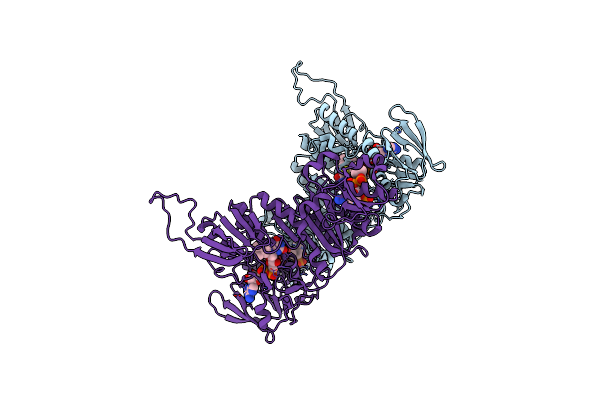 |
Structure Of The Shewanella Loihica Pv-4 Nadh-Dependent Persulfide Reductase
Organism: Shewanella loihica
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: CL, FAD, COA |
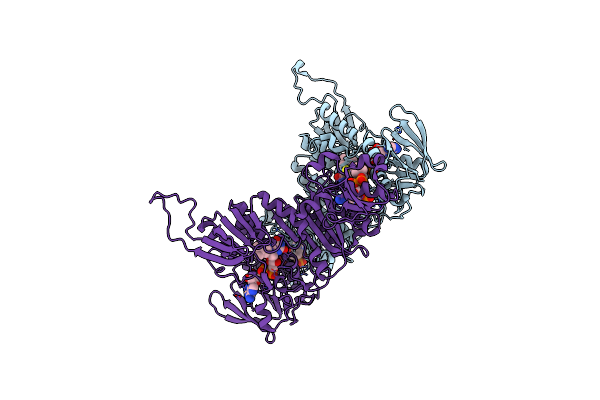 |
Structure Of The Shewanella Loihica Pv-4 Nadh-Dependent Persulfide Reductase C531S Mutant
Organism: Shewanella loihica
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: COA, CL, FAD |

