Search Count: 31
 |
Organism: Mycobacterium sp. js330
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: PEG, NA |
 |
Organism: Mycobacterium sp. js330
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: GOL, NA, CL |
 |
Organism: Mycobacterium sp. js330
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2025-01-01 Classification: HYDROLASE |
 |
Organism: Mycobacterium sp. js330
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: GOL, NA, CL, EDO, DMS |
 |
Succinate Bound Crystal Structure Of Thermus Scotoductus Sa-01 Ene-Reductase
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: SIN, FMN |
 |
Rhodococcus Ruber Alcohol Dehydrogenase Nadh Biomimetic Complex - Compound 4B
Organism: Rhodococcus ruber
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: ZN, W46, CIT, IPA |
 |
Rhodococcus Ruber Alcohol Dehydrogenase Nadh Biomimetic Complex - Compound 1A
Organism: Rhodococcus ruber
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: IPA, ZN, NA, W3O, CIT |
 |
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-07-03 Classification: FLAVOPROTEIN Ligands: FMN, W3X, IPA, NA, CL |
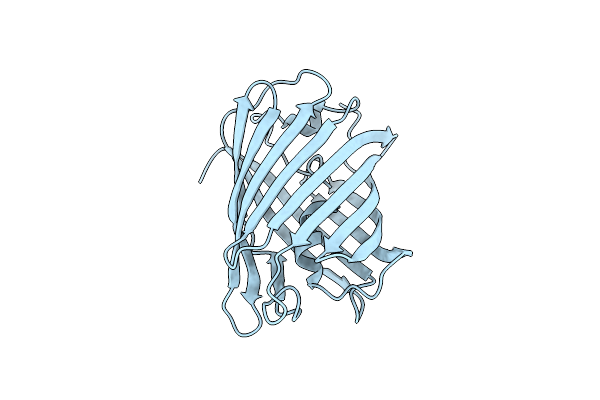 |
Organism: Galaxea fascicularis
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
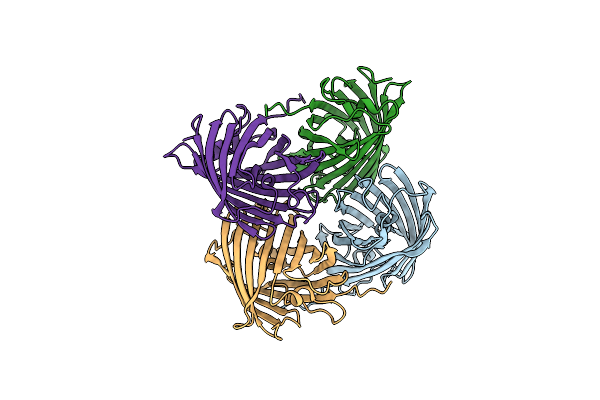
Organism: Acropora millepora
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN Ligands: CL, BR |
 |
Organism: Echinopora forskaliana
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN |
 |
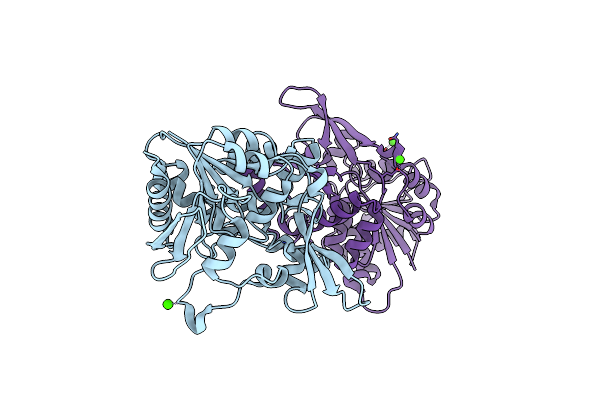
Organism: Stylophora pistillata
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN |
 |
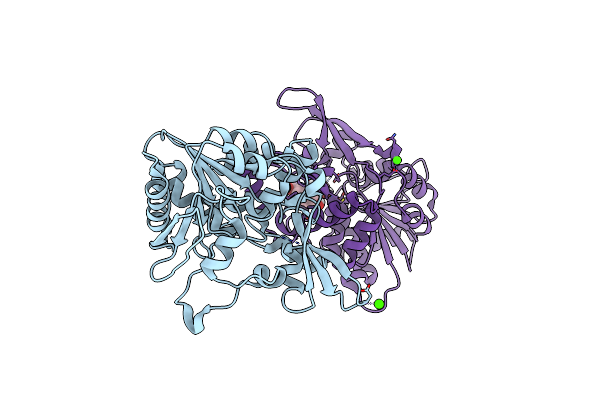
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: CA |
 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FO1, CA |
 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, GOL, CA |
 |
The Crystal Structure Of Fbia From Mycobacterium Smegmatis, Gdp And Fo Bound Form
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, FO1, CA |
 |
The Crystal Structure Of Fbia From Mycobacterium Smegmatis, Dehydro-F420-0 Bound Form
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, QMD, CA |
 |
Rational Engineering Of A Mesophilic Carbonic Anhydrase To An Extreme Halotolerant Biocatalyst
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-05-20 Classification: LYASE Ligands: ZN, GOL, NA, BOG |
 |
X-Ray Structure And Mutagenesis Studies Of The N-Isopropylammelide Isopropylaminohydrolase, Atzc
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-03-18 Classification: HYDROLASE Ligands: CL, ZN |
 |
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: FE, PEG |

