Search Count: 124
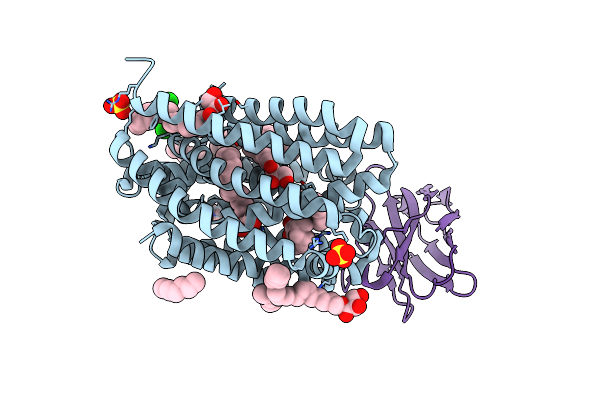 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 2.7552 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, OCT, SO4, ZN, CL |
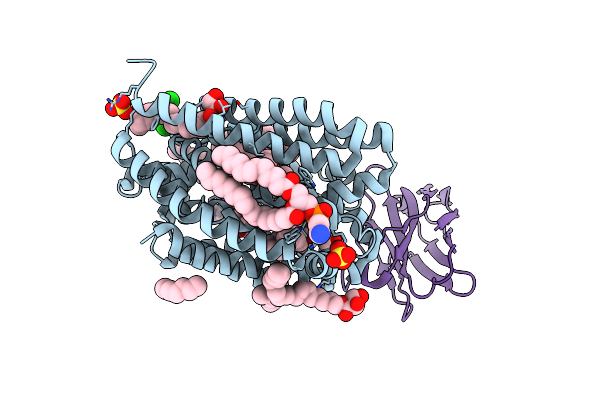 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 0.9688 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, LOP, OCT, CL, SO4, ZN |
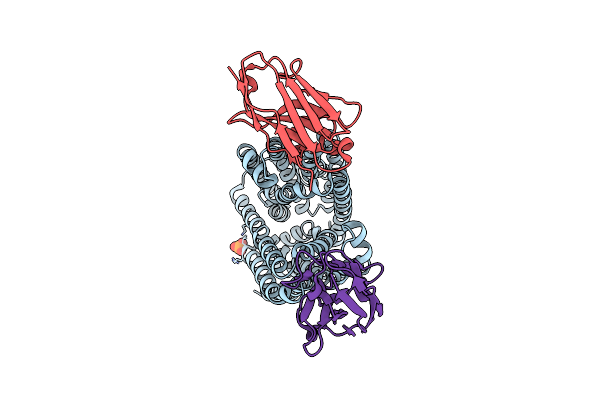 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: PO4 |
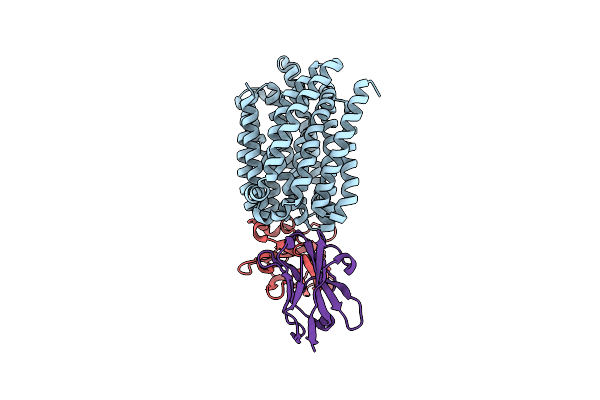 |
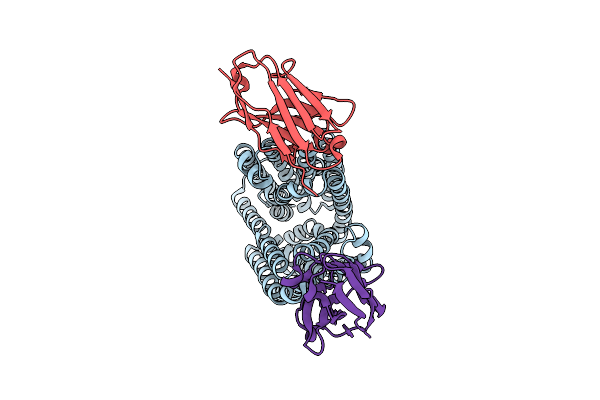
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
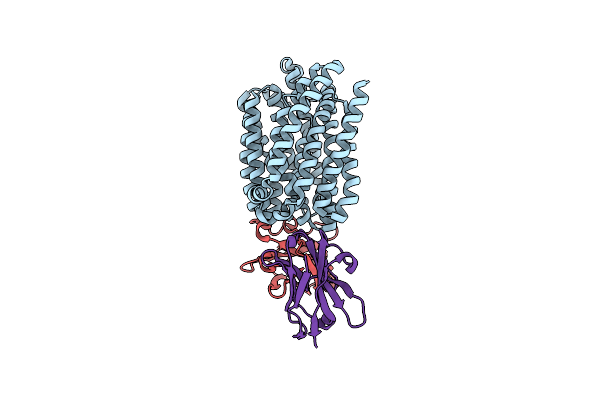
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
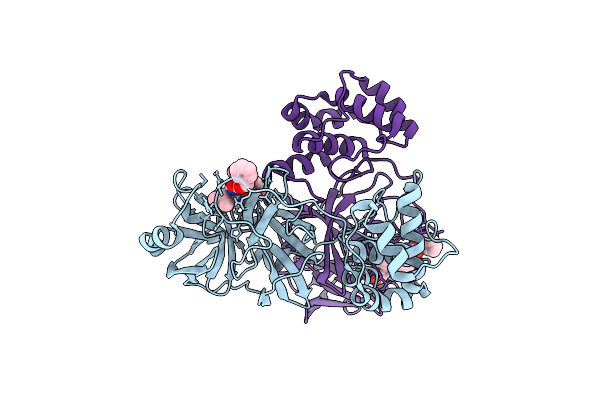
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
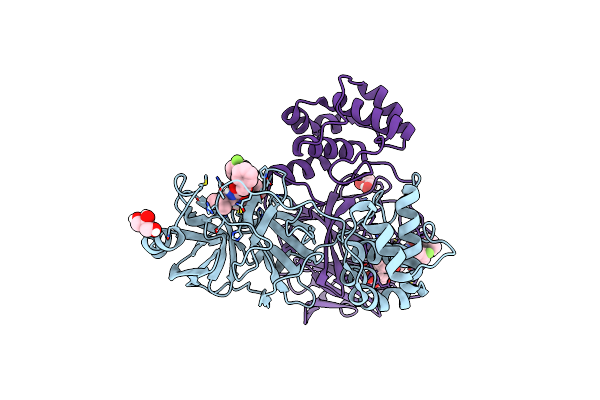
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-03-13 Classification: HYDROLASE Ligands: WTE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: WTV, GOL, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: A1ADS |
 |
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: WYR, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: MG, X5W |
 |
The Lipid Linked Oligosaccharide Polymerase Wzy And Its Regulating Co-Polymerase Wzz Form A Complex In Vivo And In Vitro
Organism: Pectobacterium atrosepticum
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN |
 |
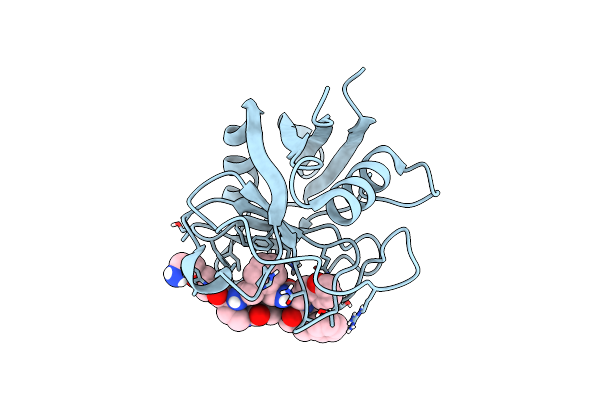
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor Jombt
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I3Q |
 |
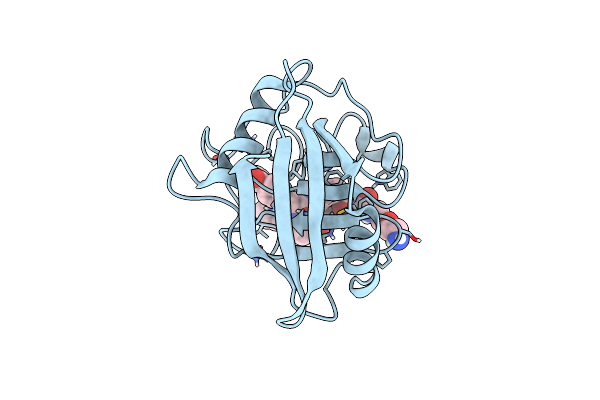
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor A26
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I4F |
 |
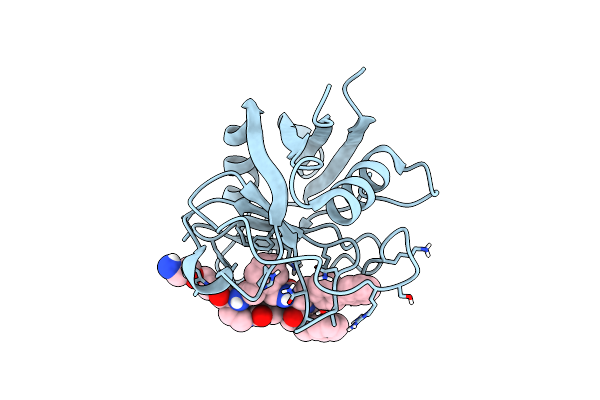
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2022-08-24 Classification: ISOMERASE/ISOMERASE inhibitor Ligands: I44 |
 |
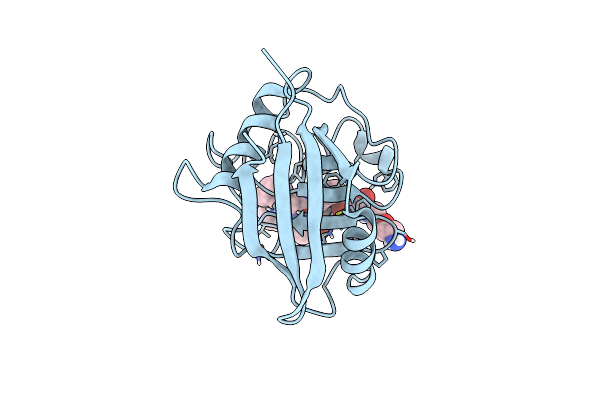
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-08-24 Classification: ISOMERASE/ISOMERASE inhibitor Ligands: I3X |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I5J |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B21
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I56 |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B23
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I5E |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B25
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I4Z |

