Search Count: 1,425
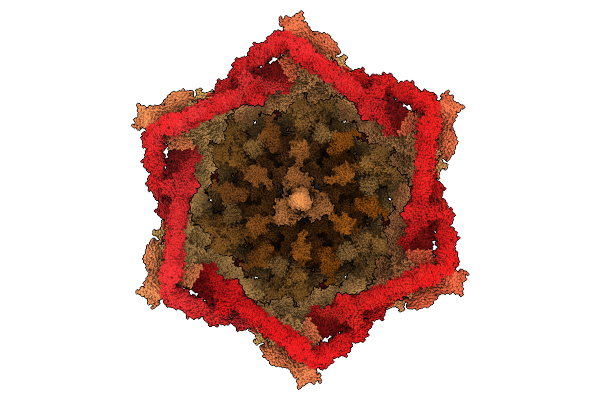 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: ZN |
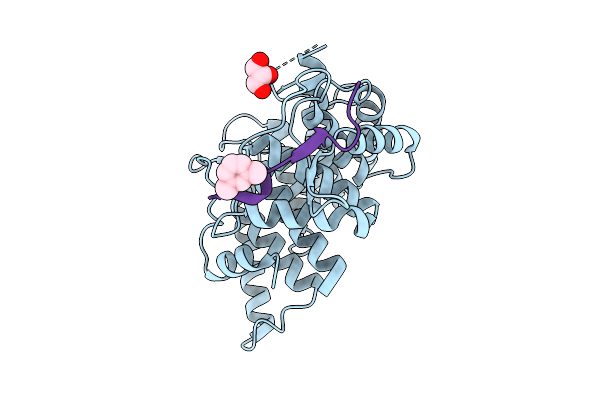 |
X-Ray Structure Analysis Of Human Complement Component 5 Te Domain In Complex With The Peptide Ra30303
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: GOL, 8VH |
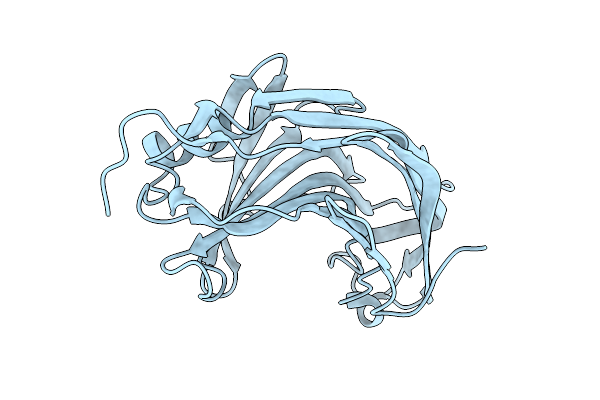 |
Crystal Structure Of The Pathogen-Secreted Apoplastic Gh12 Xyloglucan-Specific Endoglucanase Xeg1
Organism: Phytophthora sojae (strain p6497)
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE |
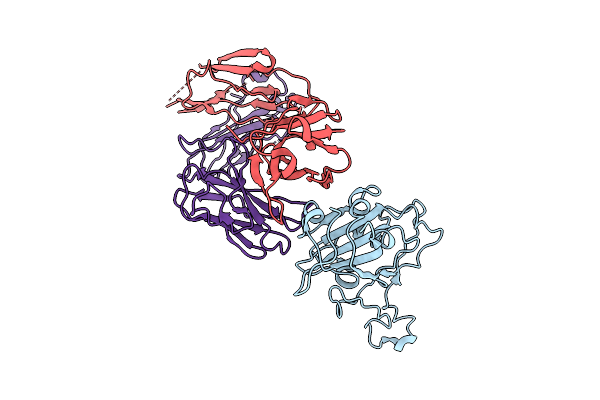 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: UNKNOWN FUNCTION |
 |
Organism: African swine fever virus (isolate tick/south africa/pretoriuskop pr4/1996)
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE |
 |
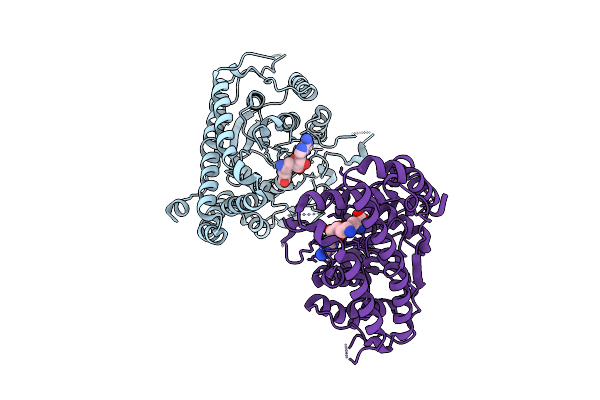
Crystal Structure Of African Swine Fever Virus Methyltransferase Ep424R In Complex With S-Adenosylmethionine
Organism: African swine fever virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: SAM |
 |
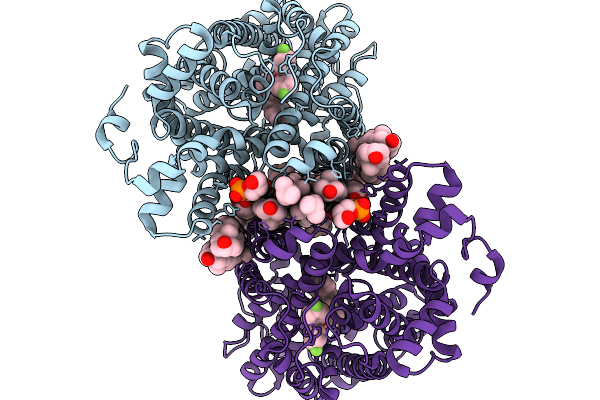
Crystal Structure Of African Swine Fever Virus Methyltransferase Ep424R In Complex With Sinefungin
Organism: African swine fever virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: SFG |
 |
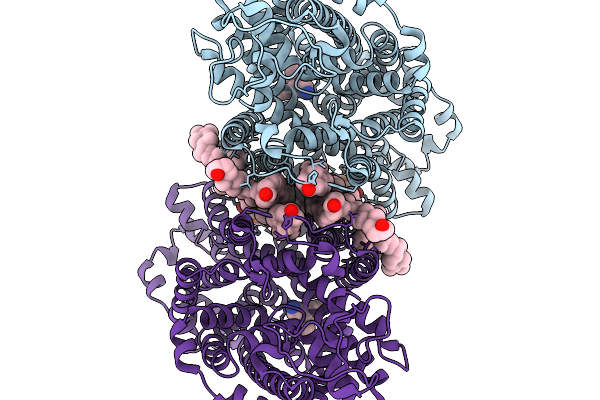
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of Vanoxerine In An Inward-Open State At Resolution Of 2.52 Angstrom
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: CLR, A1D5S, A1EFM |
 |
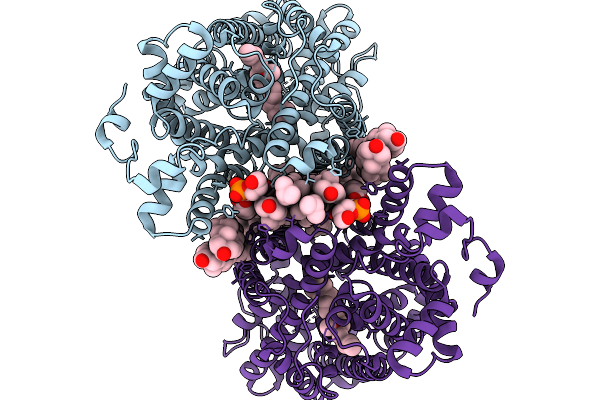
Cryo-Em Structure Of Lipid-Mediated Human Norepinephrine Transporter Net In The Presence Of Levomilnacipran In An Inward-Open State At Resolution Of 2.46 Angstrom.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: F0F, CLR, A1EFM |
 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of The F3288-0031 In An Inward-Open State At Resolution Of 3.1 Angstrom
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: A1EFR, CLR, A1EFM |
 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of The Antidepressant Vilazodone In An Inward-Open State At Resolution Of 2.44 Angstrom.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: PROTON TRANSPORT Ligands: YG7, CLR, A1EFM |
 |
Crystal Structure Of S. Aureus Protein A Bound To A Human Single-Domain Antibody
Organism: Homo sapiens, Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |

