Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
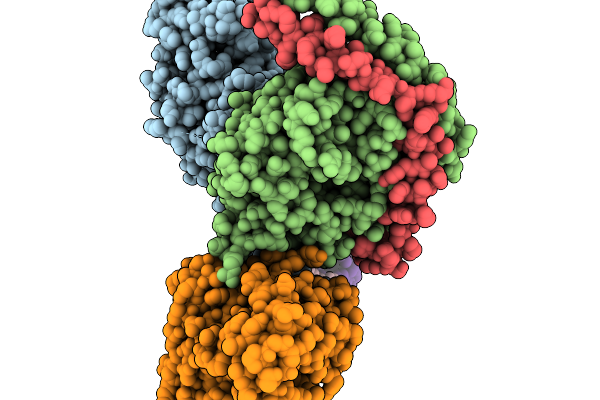 |
Positive Allosteric Modulator(Bms986187)-Bound Delta-Opioid Receptor-Gi Complex
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: A1D6B, A1D6F |
 |
Crystal Structure Of Bmp7 In Complex With 2,4-Dibromophenol Generated By Substrate Soaking
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: GOL, CL, HEM, Y8I |
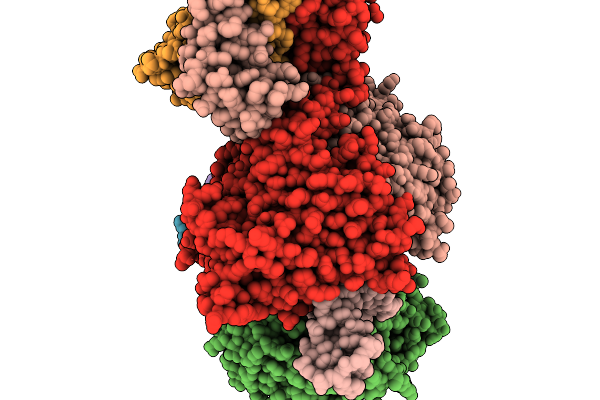 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: ATP, MG |
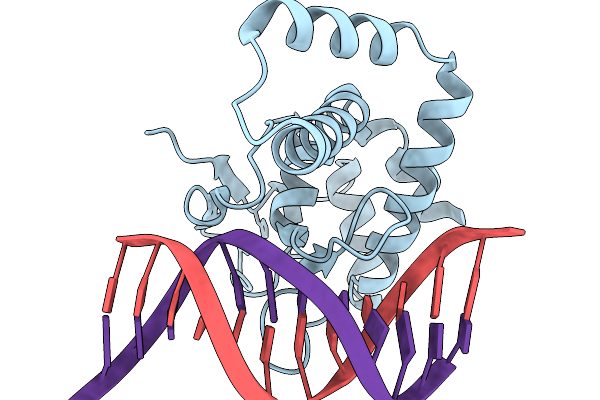 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ZN |
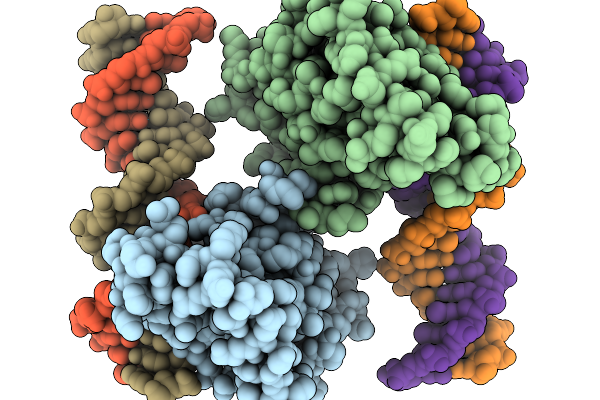 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: HEM, GOL |
 |
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: GOL, CL, HEM, Y8I |
 |
Gi-Bound Kappa Opioid Receptor In Complex With Dynorphin And Positive Allosteric Modulator Mpam-15
Organism: Homo sapiens, Rattus norvegicus, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: CLR, A1D6C |
 |
Organism: Vreelandella venusta
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: PEG |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: ATP, MG |
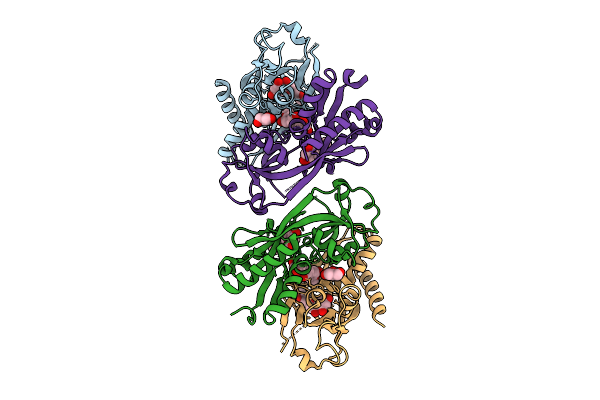 |
Crystal Structure Of Nodd-Ebd (Effector Binding Domain) In Complex With Hesperetin From Rhizobium Leguminosarum Bv. Vicae 3841
Organism: Rhizobium leguminosarum
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: GOL, 6JP |
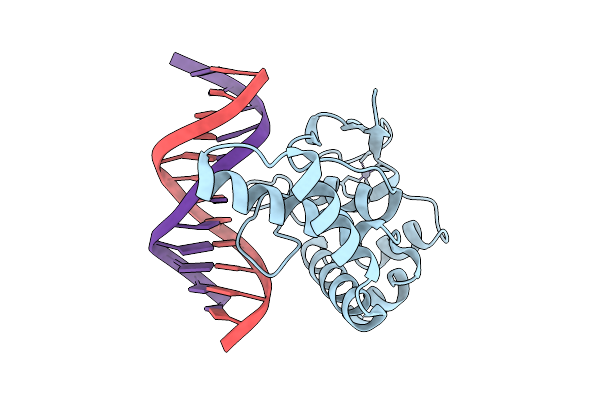 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN |
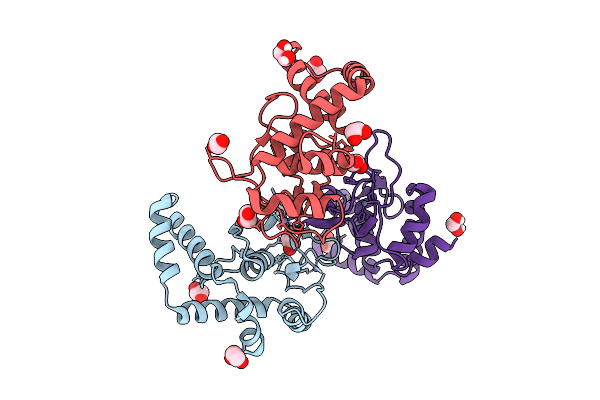 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, EDO |
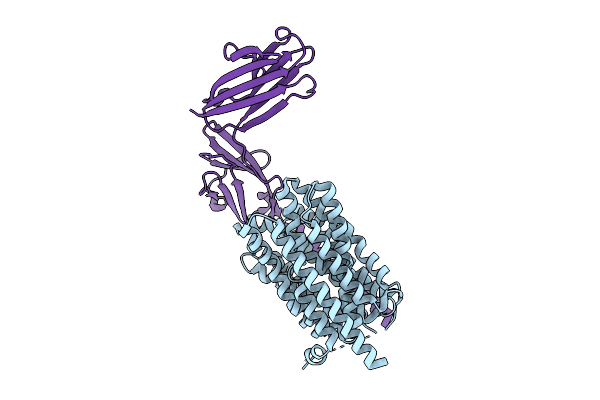 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN |
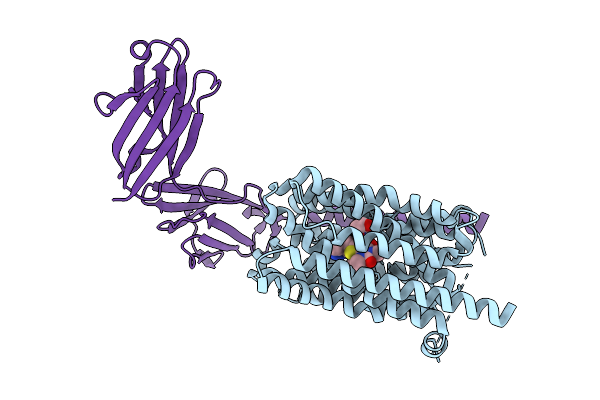 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: A1EH2 |
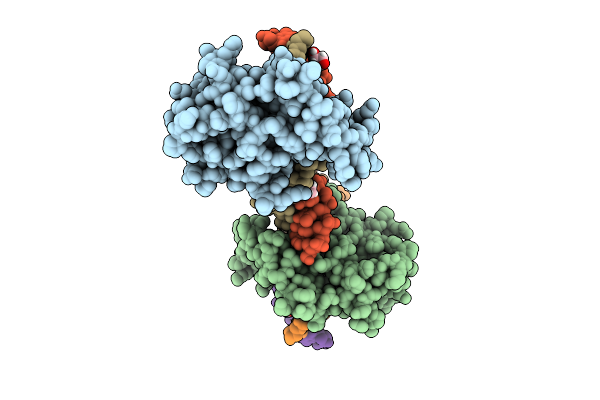 |
Crystal Structure Of Nfia In Complex With Dna Containing The Tggca(N3)Tgcca Motif
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, EDO |
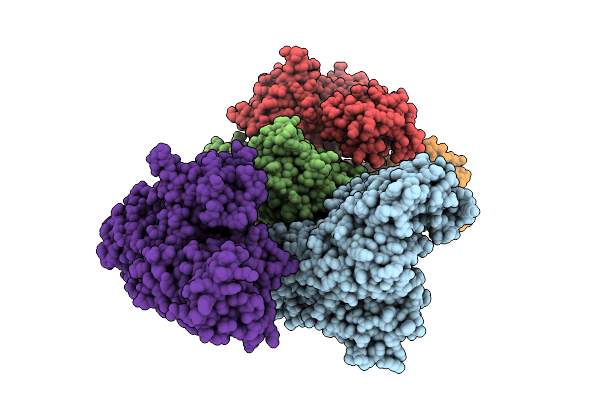 |
Crystal Structure Of A Wild-Type Tagose Isomerase (Tst4Ease Wt) From Thermotogota Bacterium
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: ISOMERASE |
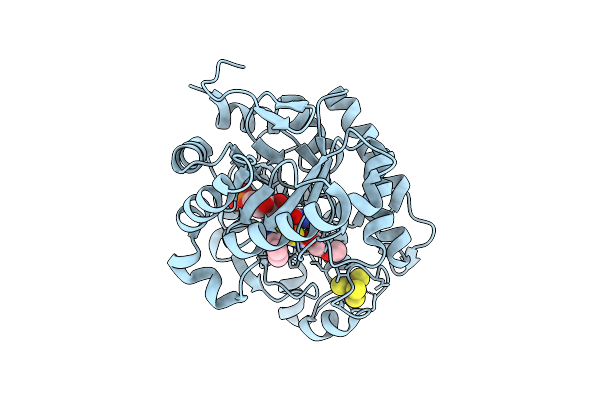 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, A1ELE, FMN |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 21
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1ELF |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 11
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1ELG |