Search Count: 40
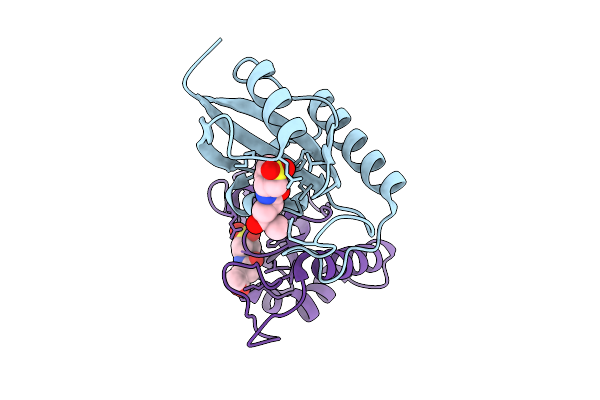 |
Human Pin1 (Peptidyl-Prolyl Cis-Trans Isomerase) Catalytic Domain Complexed With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9T |
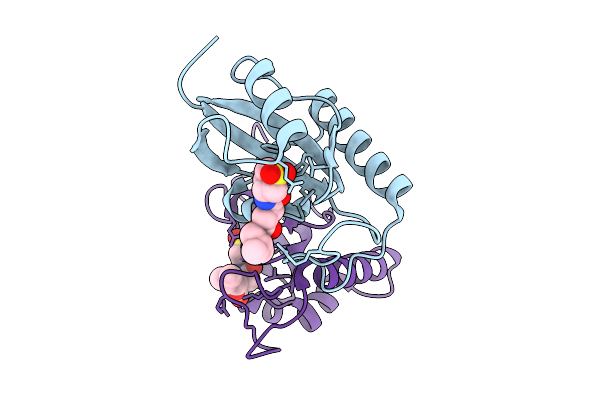 |
Human Pin1 (Peptidyl-Prolyl Cis-Trans Isomerase) Catalytic Domain In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9U |
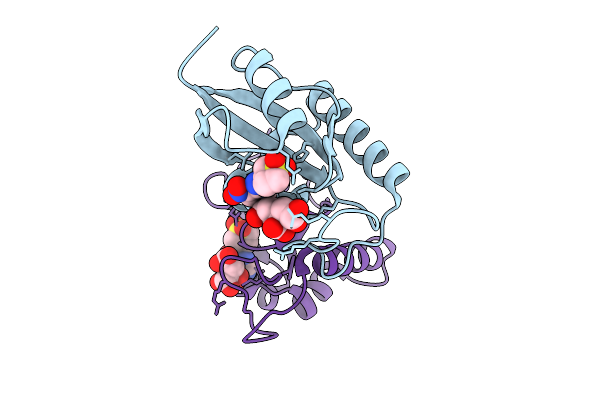 |
Crystal Structure Of Human Pin1 Catalytic Domain In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9W, CIT |
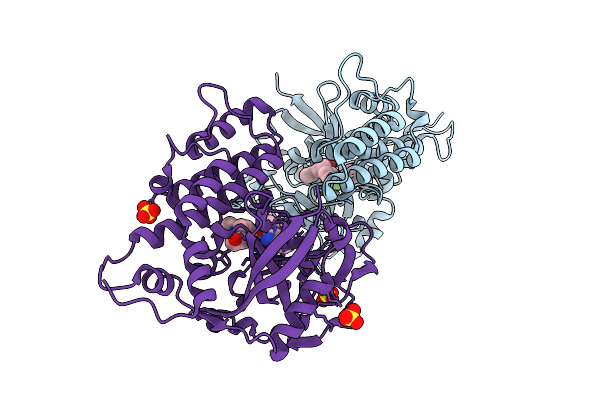 |
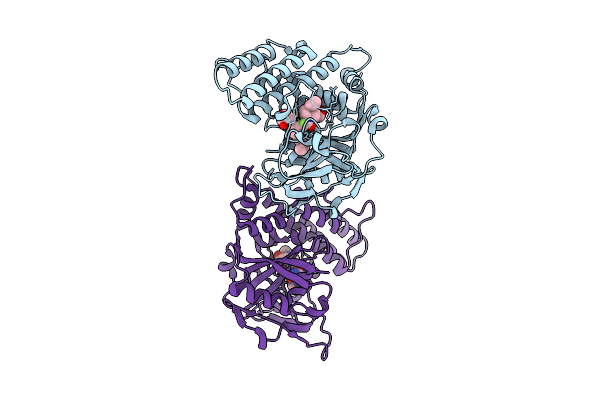
Adp-Ribosyltransferase 1 (Parp1) Catalytic Domain Bound To A Pyrimidine 2,4-Diketone Derivative Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: A1D9M, SO4 |
 |
Mutated Adp-Ribosyltransferase 2 (Parp2) Catalytic Domain Bound To A Pyrimidine 2,4-Diketone Derivative Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: GOL, A1D9M |
 |
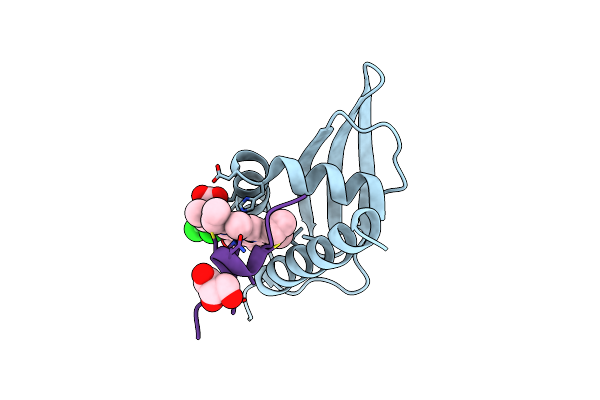
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: HEM |
 |
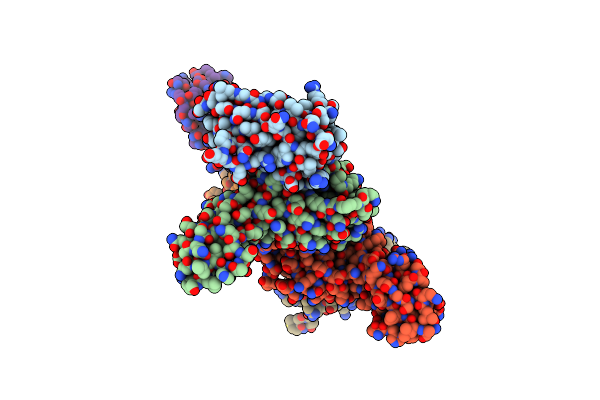
Organism: Synechocystis sp. (strain pcc 6803 / kazusa), Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: NI, CL, HEC, GOL |
 |
Organism: Vicugna pacos, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-09-04 Classification: ANTIVIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of Human Golgi Resident Glutaminyl Cyclase In Complex With (Z)-3-((1H-Benzo[D]Imidazol-5-Yl)Methylene)-4-(Piperidin-4-Yloxy)Indolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D46 |
 |
Crystal Structure Of Human Golgi Resident Glutaminyl Cyclase In Complex With (Z)-3-((1H-Benzo[D]Imidazol-5-Yl)Methylene)-4-((Tetrahydro-2H-Pyran-4-Yl)Oxy)Indolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.54 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D47 |
 |
Crystal Structure Of Human Secretory Glutaminyl Cyclase In Complex With (S)-1-(1H-Benzo[D]Imidazol-5-Yl)-5-(4-Propoxyphenyl)Imidazolidin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D49 |
 |
Crystal Structure Of Human Secretory Glutaminyl Cyclase In Complex With (Z)-3-((1H-Benzo[D]Imidazol-5-Yl)Methylene)-4-Hydroxyindolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D48 |
 |
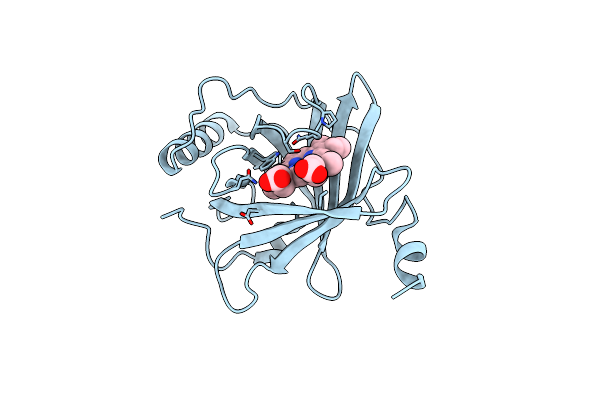
Crystal Structure Of Human Golgi Resident Glutaminyl Cyclase In Complex With (R,Z)-3-((1H-Benzo[D]Imidazol-5-Yl)Methylene)-4-((1-Acetylpyrrolidin-3-Yl)Oxy)Indolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D5C |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-12-27 Classification: PLANT PROTEIN Ligands: HEM |
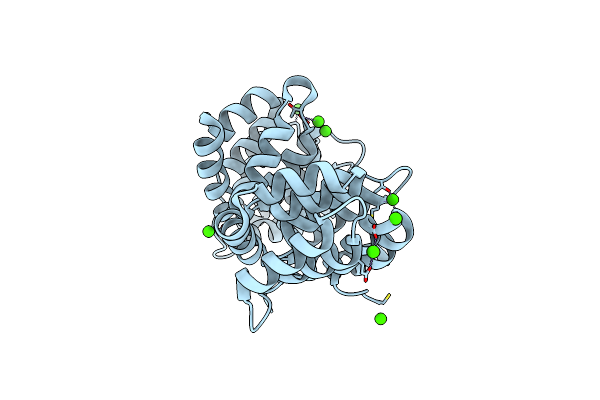 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-11-01 Classification: LIPID BINDING PROTEIN Ligands: CA |
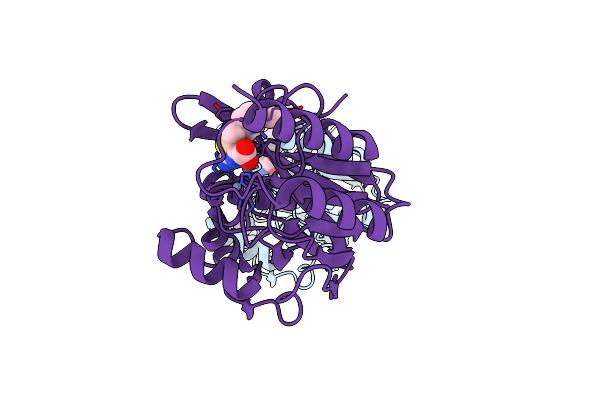 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-(4-Methoxybenzyl)Thiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 4YJ |
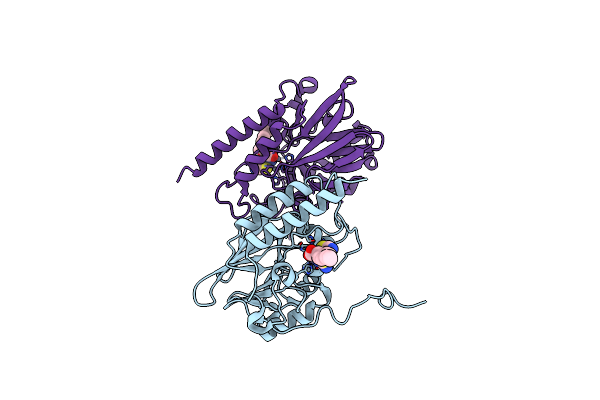 |
Crystal Structure Of B3 L1 Mbl In Complex With 2-Amino-5-(4-Propylbenzyl)Thiazole-4-Carboxylic Acid
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 51I, SCN |
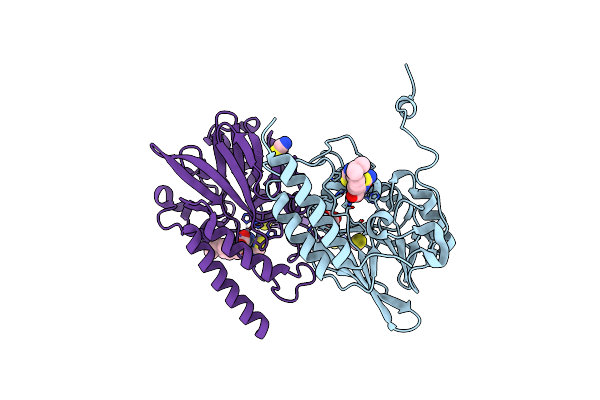 |
Crystal Structure Of B3 L1 Mbl In Complex With 2-Amino-5-(4-Isopropylbenzyl)Thiazole-4-Carboxylic Acid
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5A5, SCN, GOL |
 |
Crystal Structure Of B2 Sfh-I Mbl In Complex With 2-Amino-5-(4-(But-3-En-1-Yloxy)Benzyl)Thiazole-4-Carboxylic Acid
Organism: Serratia fonticola
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5I6, EOH |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-Isobutylthiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5IY, GOL |

