Search Count: 2,585
 |
Adp-Ribosyltransferase 1 (Parp1) Catalytic Domain Bound To A Pyrimidine 2,4-Diketone Derivative Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: A1D9M, SO4 |
 |
Mutated Adp-Ribosyltransferase 2 (Parp2) Catalytic Domain Bound To A Pyrimidine 2,4-Diketone Derivative Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: GOL, A1D9M |
 |
Neuraminidase Of A/Red Knot/Delaware Bay/310/2016 H10N4 In Complex With Cav-F6 Fab
Organism: Influenza a virus (a/red knot/delaware bay/310/2016(h10n4)), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CA |
 |
Organism: Homo sapiens, Influenza a virus (a/california/04/2009(h1n1))
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CA |
 |
Neuraminidase Of A/Switzerland/9715293/2013 H3N2 In Complex With Cav-F34 Fab
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CA |
 |
Neuraminidase Of A/Switzerland/9715293/2013 H3N2 In Complex With Cav-F6 Fab
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CA |
 |
Neuraminidase Of A/Switzerland/9715293/2013 H3N2 In Complex With Cav-F6-R54S Fab
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CA |
 |
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: NAG, CA |
 |
Neuraminidase Of A/Dairy Cow/Minnesota/24_016288-003/2024 (Dcmn24 N1) In Complex With Cav-F6 Fab
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CA |
 |
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: NAG, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: ZN, MG, A1EBE |
 |
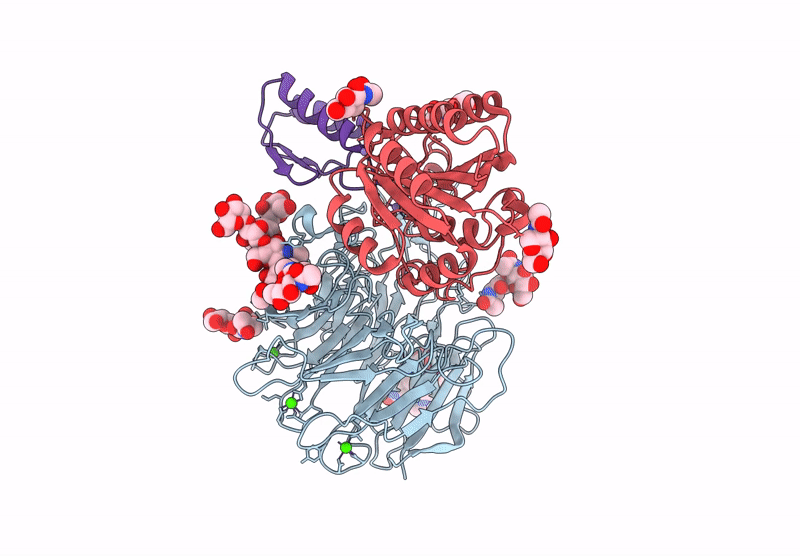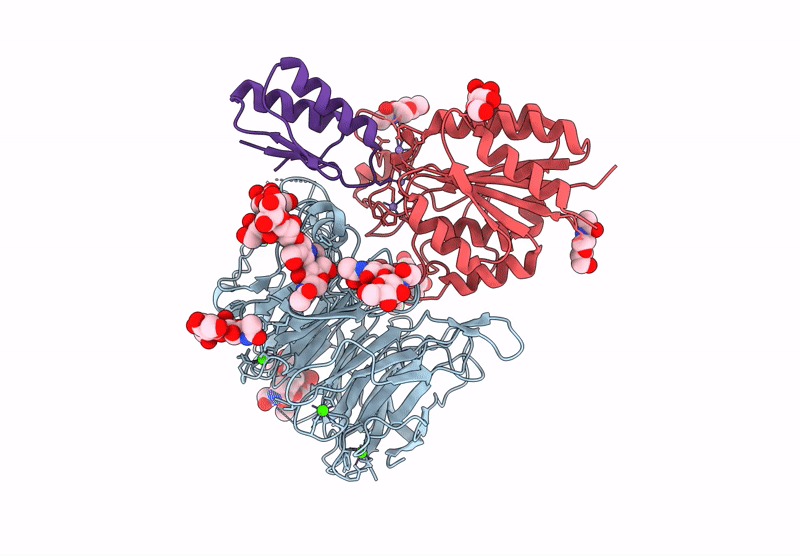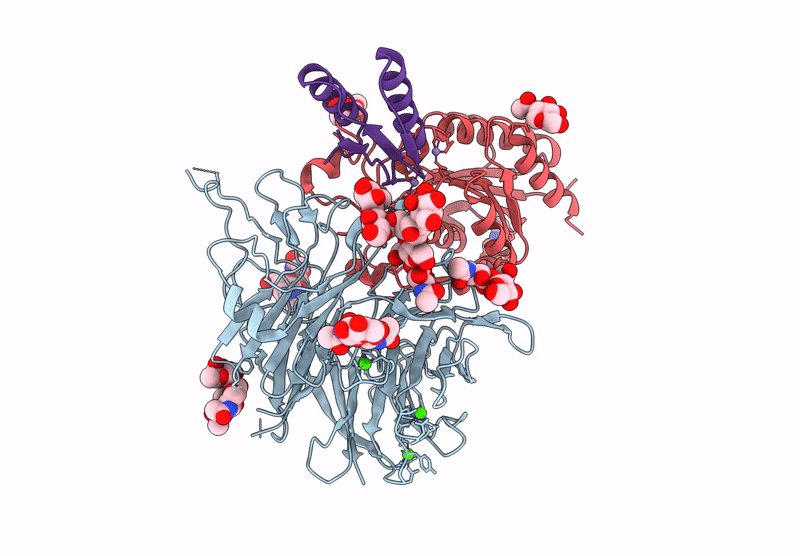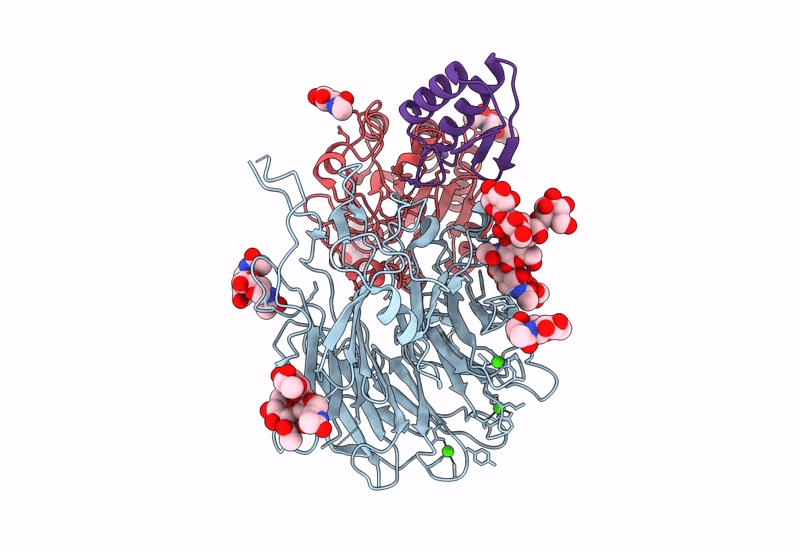
Cryo-Em Structure Of Alpha5Beta1 Integrin In Complex With Neonectin Candidate 2
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: SIGNALING PROTEIN Ligands: CA, NAG, MN |
 |
Cryo-Em Structure Of Alpha5Beta1 Integrin In Complex With Neonectin Candidate 2, Open Conformation
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: SIGNALING PROTEIN Ligands: CA, NAG, MN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: A1ELJ, ZN, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: A1ELC, ZN, MG, SO4 |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-06-04 Classification: GENE REGULATION Ligands: O6E |
 |
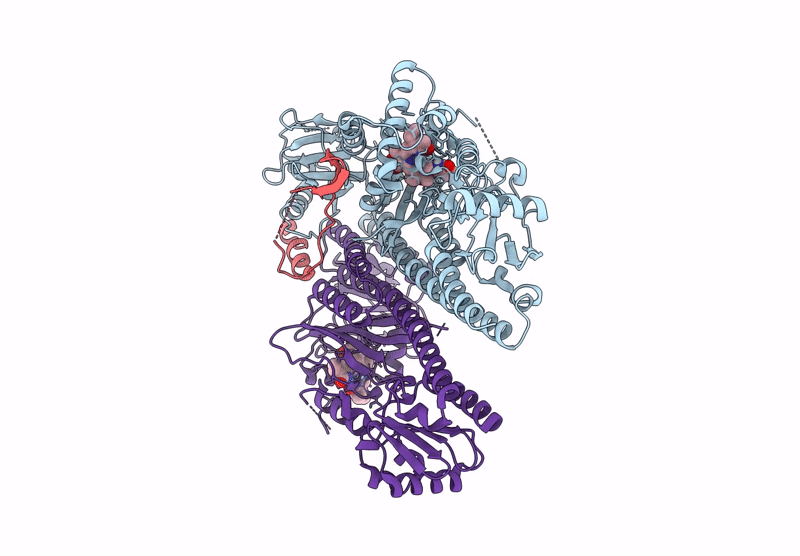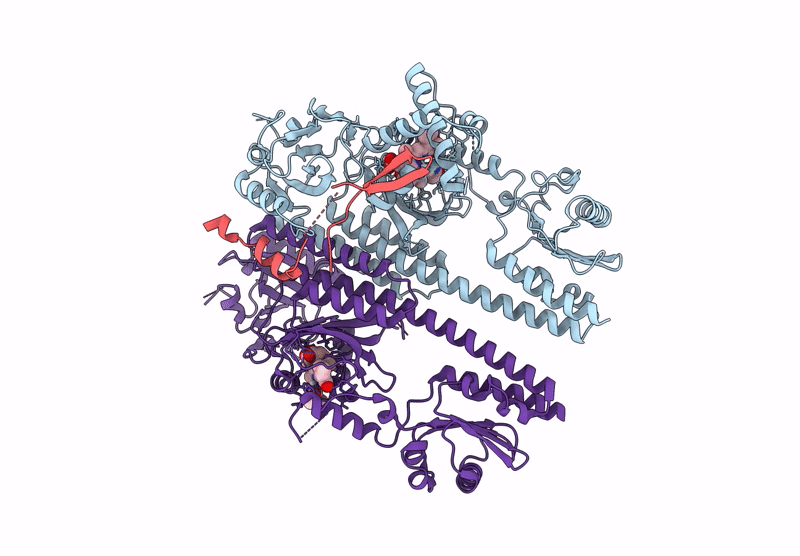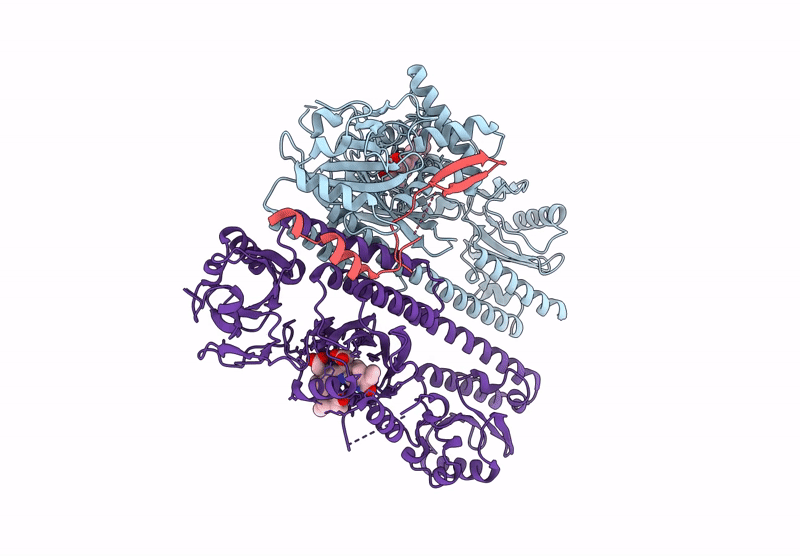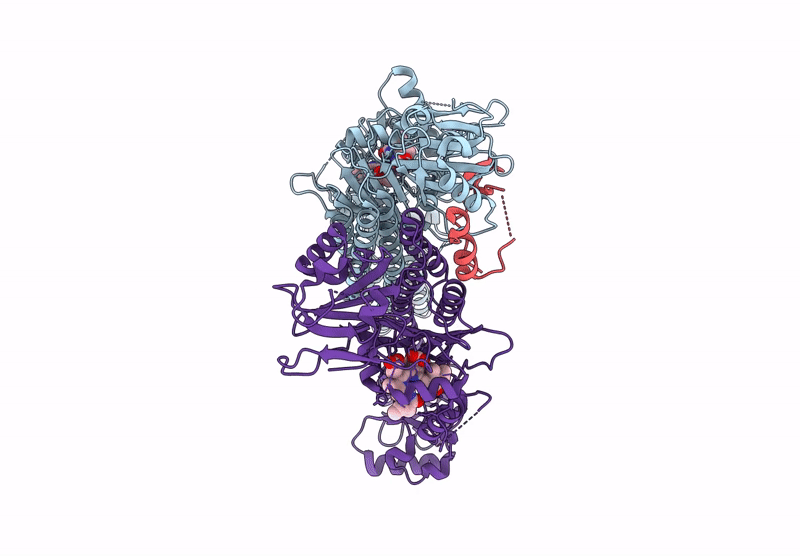
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-06-04 Classification: GENE REGULATION Ligands: O6E |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-06-04 Classification: GENE REGULATION Ligands: O6E |
 |
Organism: Streptomyces cinnamoneus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: FE2, AKG, GOL |

