Search Count: 16
 |
 |
Organism: Asplenium bulbiferum subsp. bulbiferum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: LYASE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-11-16 Classification: FLUORESCENT PROTEIN Ligands: CA, GOL, TLA |
 |
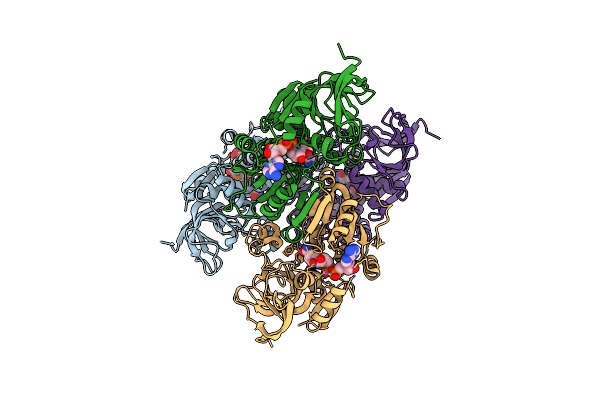
Crystal Structure Of Mtb Pks13-Te In Complex With Inhibitor Coumestan Derivative 8
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: 7IJ |
 |
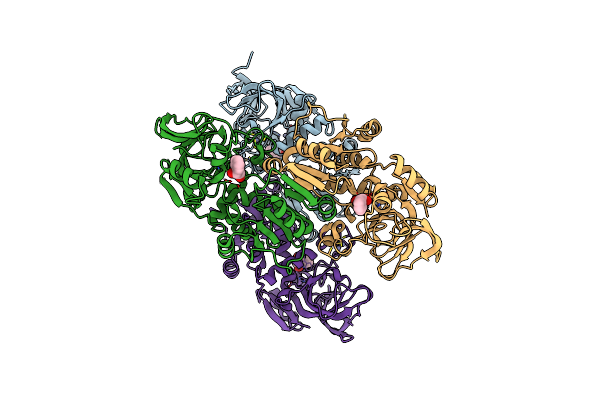
Crystal Structure Of (R)-Carbonyl Reductase From Candida Parapsilosis In Complex With Nad
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
 |
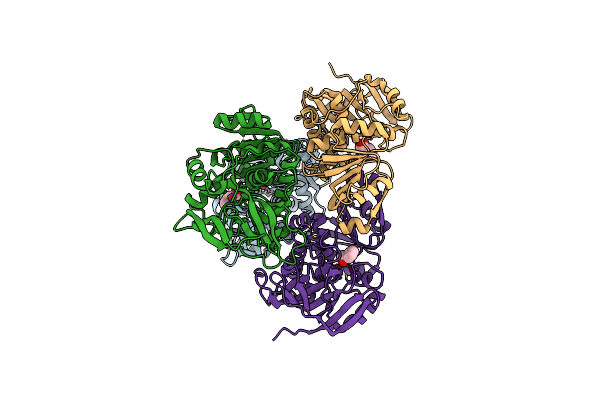
Crystal Structure Of (R)-Carbonyl Reductase From Candida Parapsilosis In Complex With (R)-1-Phenyl-1,2-Ethanediol
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: FEH, ZN |
 |
Crystal Structure Of (R)-Carbonyl Reductase H49A Mutant From Candida Parapsilosis In Complex With 2-Hydroxyacetophenone
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: HXT, ZN |
 |
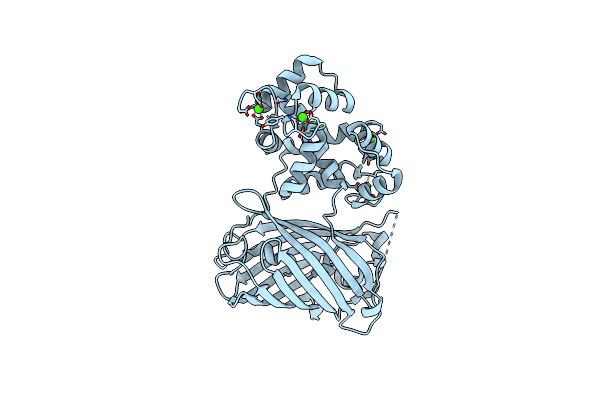
Organism: Gallus gallus, Aequorea victoria, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-20 Classification: FLUORESCENT PROTEIN Ligands: CA |
 |
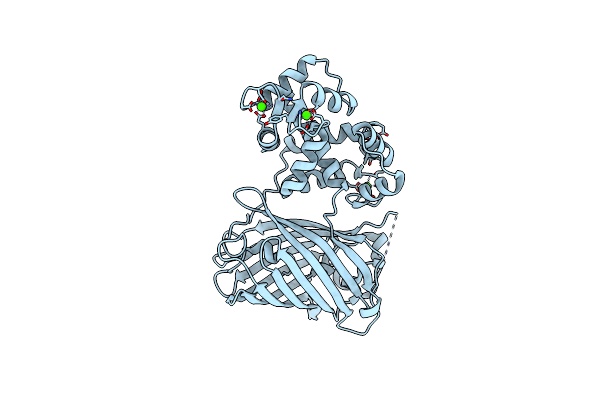
Organism: Gallus gallus, Aequorea victoria, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-06-20 Classification: FLUORESCENT PROTEIN Ligands: CA |
 |
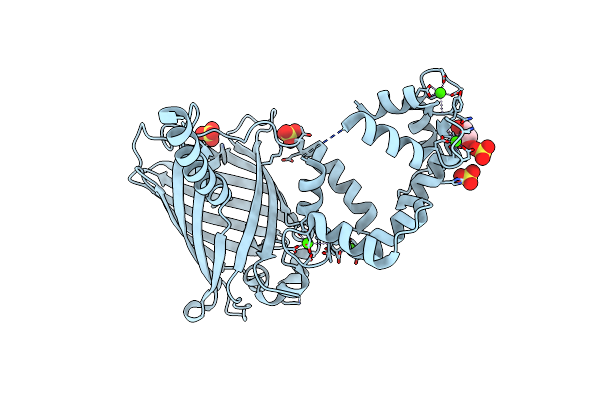
Organism: Gallus gallus, Aequorea victoria, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-06-20 Classification: FLUORESCENT PROTEIN Ligands: CA |
 |
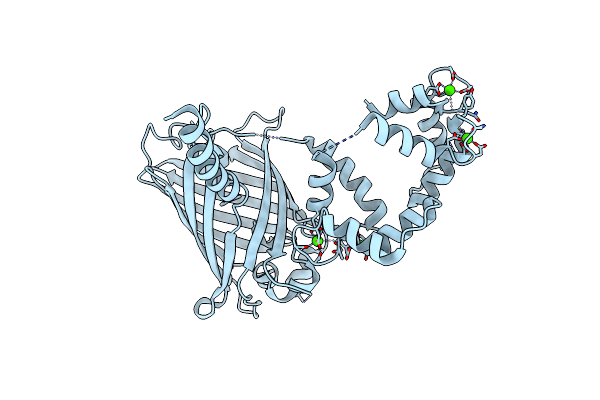
Organism: Gallus gallus, Aequorea victoria, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-20 Classification: FLUORESCENT PROTEIN Ligands: CA, SO4, GOL |
 |
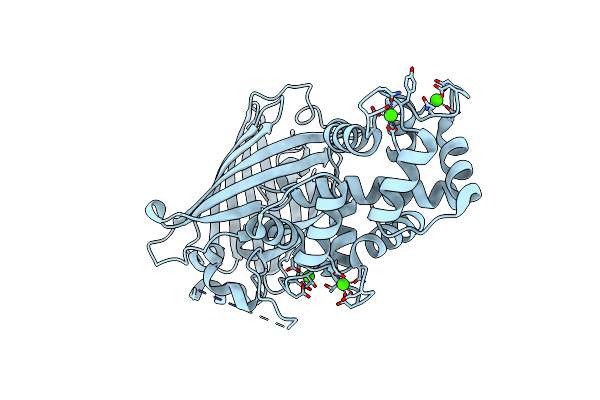
Organism: Gallus gallus, Aequorea victoria, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-06-20 Classification: FLUORESCENT PROTEIN Ligands: CA |
 |
Organism: Gallus gallus, Aequorea victoria, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-20 Classification: FLUORESCENT PROTEIN Ligands: CA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2009-12-08 Classification: CHAPERONE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2009-12-08 Classification: CHAPERONE Ligands: BME, CL |
 |
The Structure Of Fis Mutant Pro61Ala Illustrates That The Kink Within The Long Alpha-Helix Is Not Due To The Presence Of The Proline Residue
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-02-14 Classification: DNA BINDING PROTEIN |

