Search Count: 440
 |
Organism: Homo sapiens, Lama glama, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, SO4 |
 |
Organism: Marinobacter sp. dsm 11874
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: 1GP |
 |
Organism: Marinobacter sp. dsm 11874
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: G3P |
 |
Organism: Phaeobacter sp. med193
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: 1GP |
 |
Organism: Phaeobacter sp. med193
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: G3P |
 |
Organism: Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: HYDROLASE |
 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Esr (L7-D1), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Esr (L11-C6), Bound To Ethametsulfuron-Methyl
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: RXF |
 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Csr (L4.2-20), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Csr (L4.2-20), Bound To Chlorsulfuron
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: 1CS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW9 |
 |
Crystal Structure Of The Acinetobacter Baumannii Lysr Family Regulator Acer Dna-Binding Domain (P321)
Organism: Acinetobacter baumannii (strain atcc 17978 / dsm 105126 / cip 53.77 / lmg 1025 / ncdc kc755 / 5377)
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DE NOVO PROTEIN Ligands: GOL |
 |
Organism: Weissella ceti
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: PG4 |
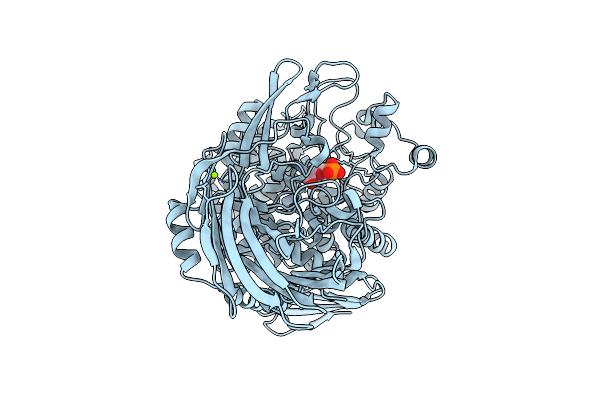 |
Crystal Structure Of Trehalose-6-Phosphate Phosphorylase From Weissella Ceti In Complex With Beta-Glc1P
Organism: Weissella ceti
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: XGP, MG |

