Search Count: 1,898
 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PLANT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: CLR, NAG |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: 8K3 |
 |
Organism: Serratia marcescens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: ANTIVIRAL PROTEIN Ligands: GTP |
 |
Structure Of The Sweet Receptor Bound To Sucralose In The Loose State, Extracellular Domain
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
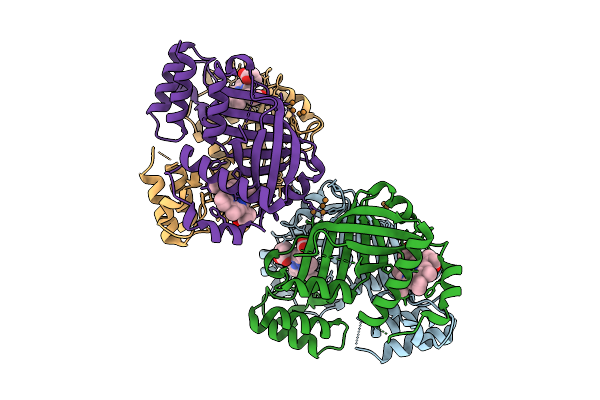 |
Organism: Streptomyces parvus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, CU |
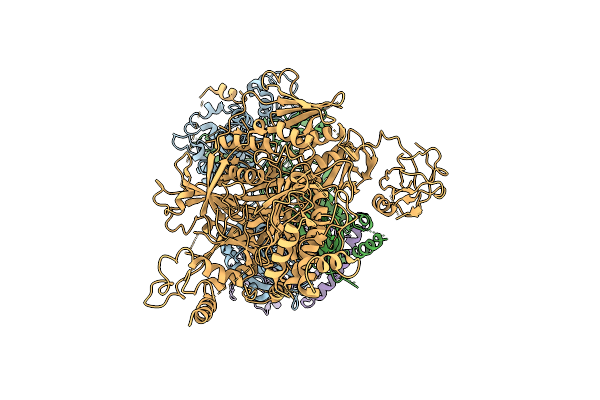 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
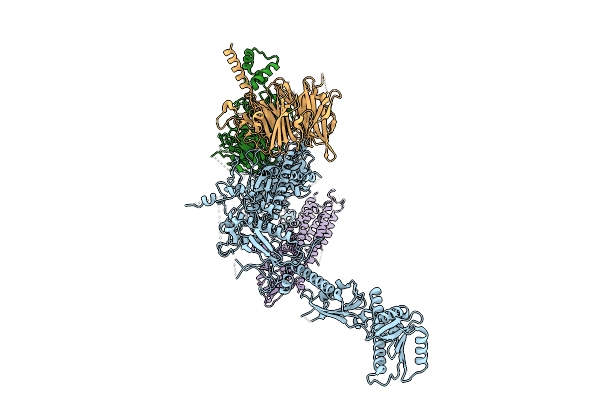 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
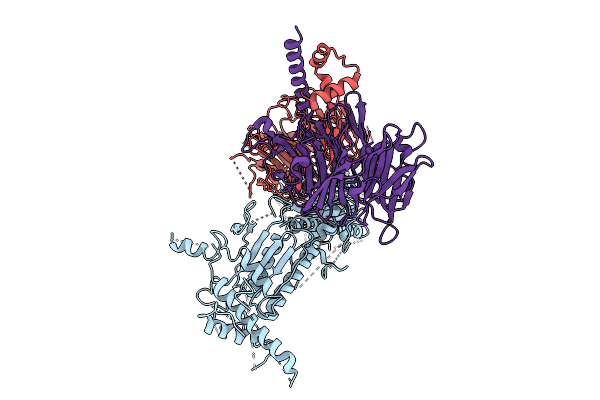 |
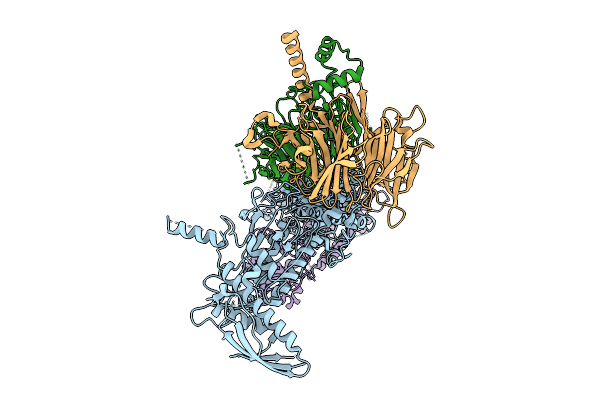
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
 |
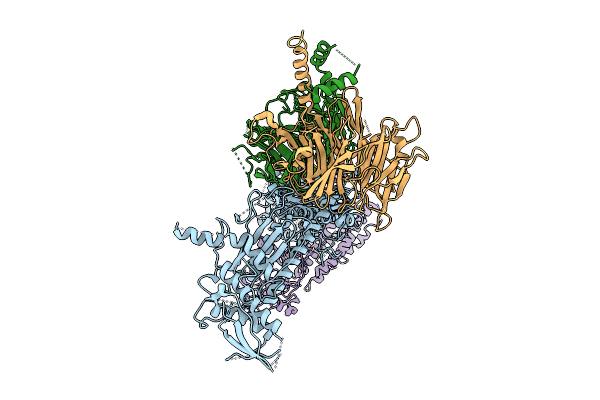
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
 |
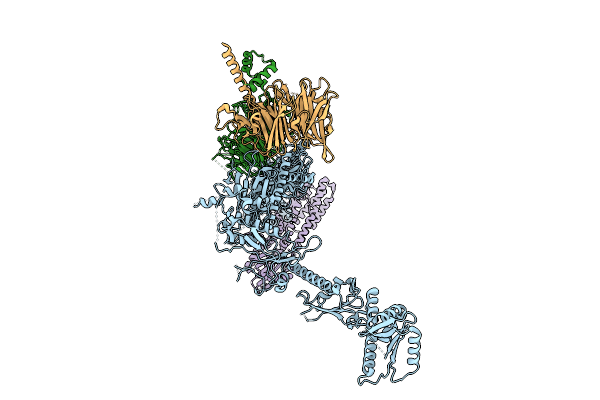
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
 |
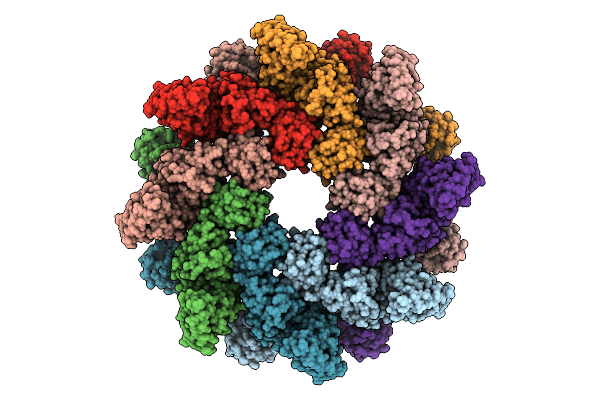
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTEIN FIBRIL |
 |
Crystal Structure Of Anae In Apo Form, A Fungal Gdsl-Acetylesterase With Acetylcholinesterase-Like Activity From Aspergillus Niger
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: MES |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: ACT, GOL |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE |

