Search Count: 13
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-08-24 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-08-24 Classification: HYDROLASE Ligands: EIB |
 |
Crystal Structure Of Mbp Fused Activation-Induced Cytidine Deaminase (Aid) In Complex With Cacodylic Acid
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: ZN, CAC, CA |
 |
Crystal Structure Of Mbp Fused Activation-Induced Cytidine Deaminase (Aid) In Complex With Dcmp
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-08-16 Classification: HYDROLASE/DNA Ligands: ZN, DCM, CA, GOL |
 |
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.61 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: ZN |
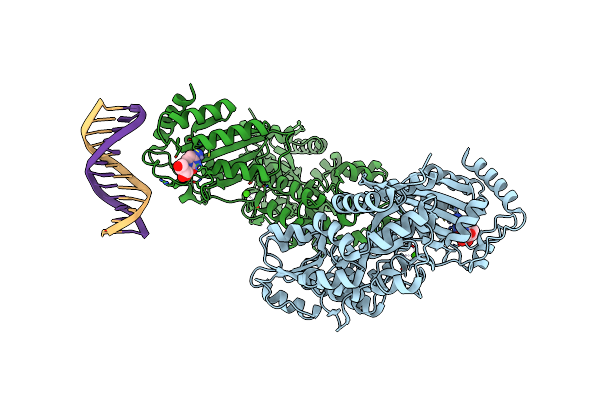 |
Crystal Structure Of Mbp Fused Activation-Induced Cytidine Deaminase (Aid) In Complex With Cytidine
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2017-08-16 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, CTN, CA |
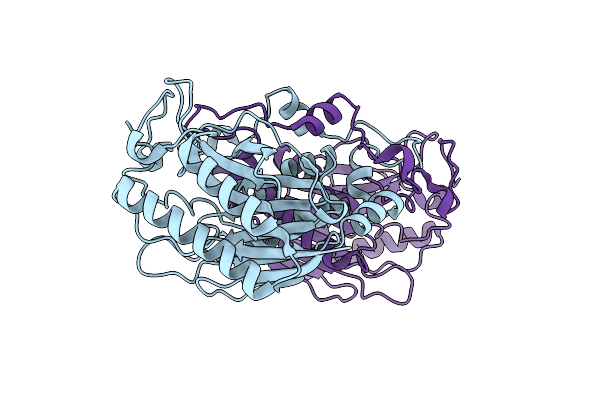 |
Helicobacter Pylori Formamidase Amif Contains A Fine-Tuned Cysteine-Glutamate-Lysine Catalytic Triad
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-02-13 Classification: HYDROLASE |
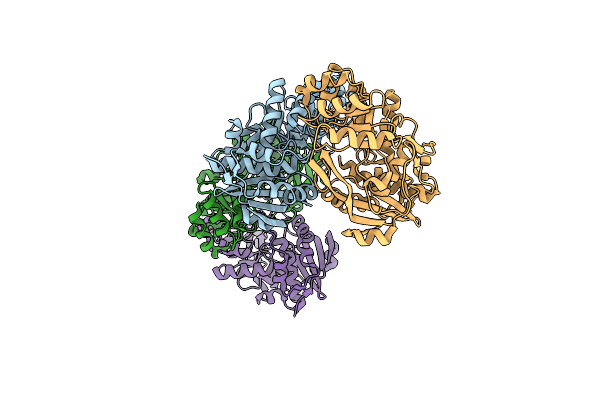 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-04-11 Classification: ISOMERASE |
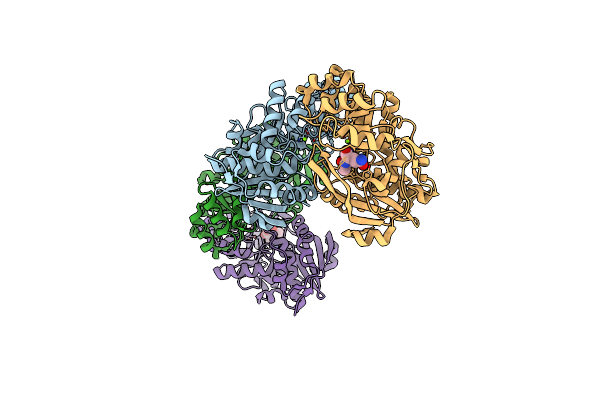 |
The Mutant A68C-D72C-Nlq Of Deinococcus Radiodurans Nacylamino Acid Racemase
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-04-11 Classification: ISOMERASE Ligands: MG, NLQ |
 |
The Mutant E149C-A182C Of Deinococcus Radiodurans N-Acylamino Acid Racemase
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-04-11 Classification: ISOMERASE |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-04-11 Classification: ISOMERASE |
 |
The Mutant A302C Of Agrobacterium Radiobacter N-Carbamoyl-D-Amino-Acid Amidohydrolase
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-04-11 Classification: HYDROLASE |
 |
The Mutant A222C Of Agrobacterium Radiobacter N-Carbamoyl-D-Amino Acid Amidohydrolase
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-04-11 Classification: HYDROLASE |

