Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Norovirus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR Ligands: SO4, NA |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: METAL BINDING PROTEIN |
 |
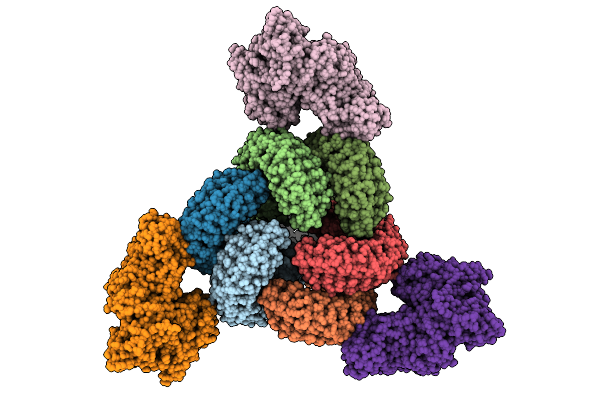
Organism: Alteromonas macleodii
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: GOL, FE, CA, MN, MG |
 |
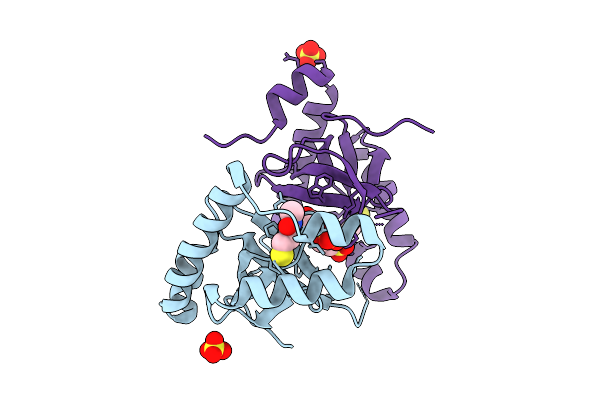
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
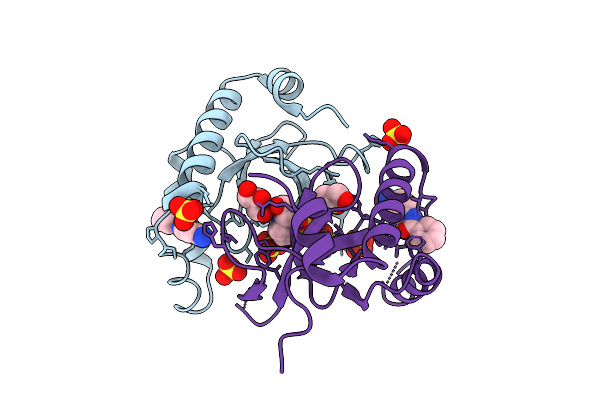
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: SO4, A1EF6, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: 6Y3, SO4, GOL, EPE |
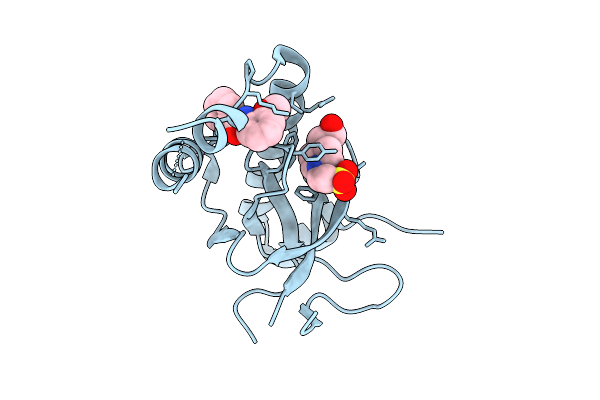 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: EPE, A1EF7 |
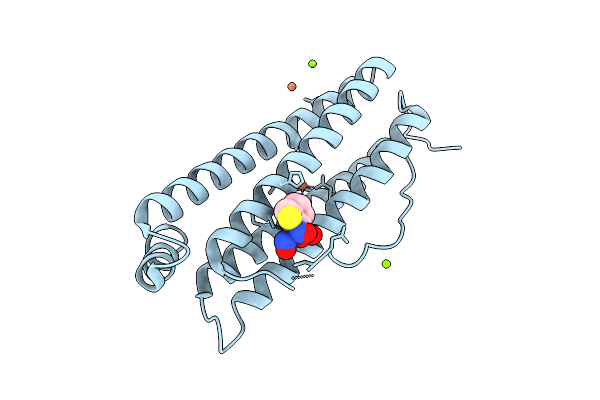 |
Crystal Structure Of Human Heavy Chain Ferritin Binding With Dinitrosyl Iron Complex Modificated By Phenylboronic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL, NO, A1EKS, H2S |
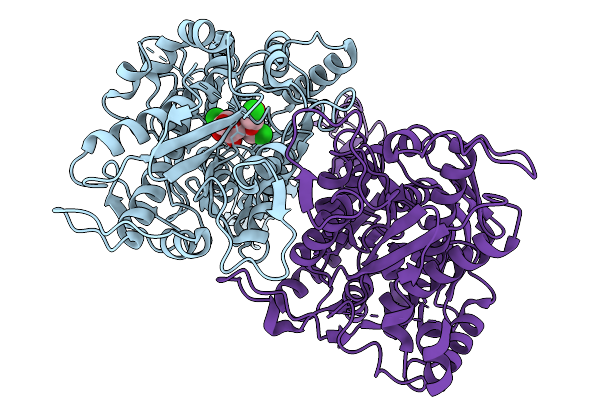 |
Structure Of The Sweet Receptor Bound To Sucralose In The Loose State, Extracellular Domain
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
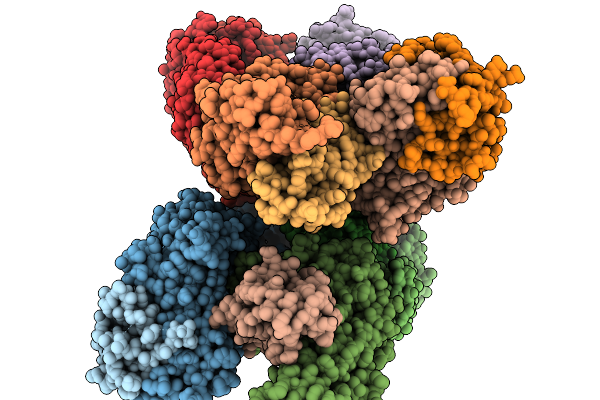 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: IMMUNE SYSTEM Ligands: GOL, PEG |
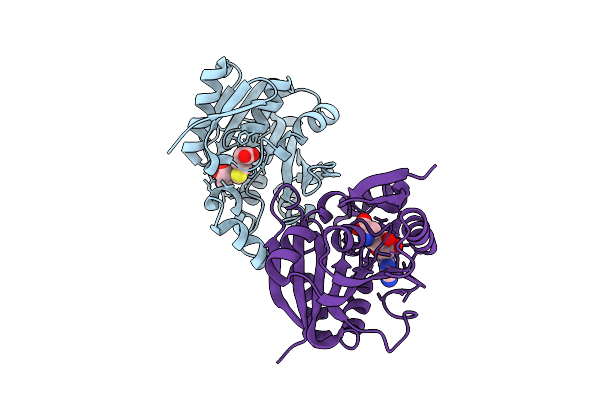 |
Crystal Structure Of The Carbamoyl N-Methyltransferase Asc-Orf2 Complexed With Sah
Organism: Amycolatopsis alba dsm 44262
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH |
 |
Organism: Mycoplasma pneumoniae (strain atcc 29342 / m129 / subtype 1)
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, MG |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOSOLIC PROTEIN Ligands: AG |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Leptinotarsa decemlineata
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE |
 |
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: ANTITUMOR PROTEIN |
 |
Organism: Bacterium hr29
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE |
 |
Crystal Structure Of A Bifunctional 3-Hexulose-6-Phosphate Synthase/6-Phospho-3-Hexuloisomerase
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: ISOMERASE Ligands: MG, SO4 |