Search Count: 220
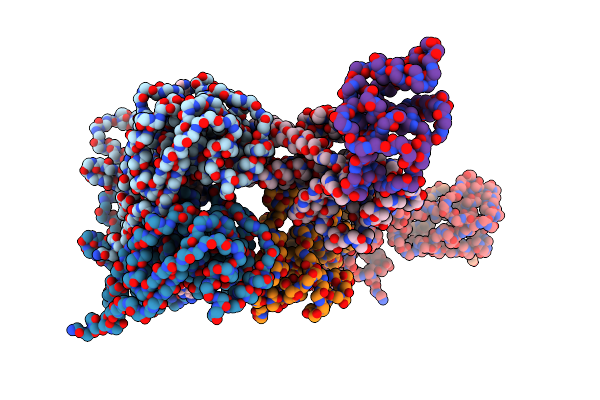 |
Intracellular Cryo-Tomography Structure Of Ebov Nucleocapsid At 8.9 Angstrom
Organism: Ebola virus - mayinga, zaire, 1976, Zaire
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: VIRAL PROTEIN |
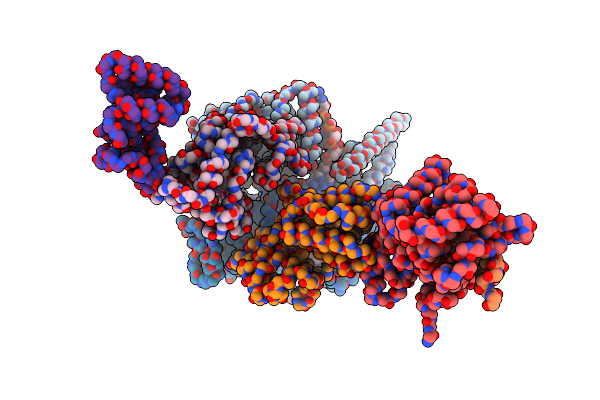 |
In-Virion Structure Of Ebola Virus Nucleocapsid-Like Assemblies From Recombinant Virus-Like Particles (Nucleoprotein, Vp24,Vp35,Vp40)
Organism: Ebola virus - mayinga, zaire, 1976, Zaire
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: VIRAL PROTEIN/RNA |
 |
Cryoem Structure Of A C7-Symmetrical Groel7-Groes7 Cage In Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel7-Groes7 Cage With Encapsulated Disordered Substrate Metk In The Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel7-Groes7 Cage With Encapsulated Ordered Substrate Metk In The Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Structure Average Of Groel14 Complexes Found In The Cytosol Of Escherichia Coli Overexpressing Groel Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
In Situ Structure Average Of Groel14-Groes14 Complexes In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Cryoem Structure Of A Groel14-Groes7 Complex In Presence Of Adp-Befx With Wide Groel7 Trans Ring Conformation
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel14-Groes7 Complex In Presence Of Adp-Befx With Narrow Groel7 Trans Ring Conformation
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
In Situ Structure Average Of Groel14-Groes7 Complexes With Wide Groel7 Trans Ring Conformation In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
In Situ Structure Average Of Groel14-Groes7 Complexes With Narrow Groel7 Trans Ring Conformation In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: VIRAL PROTEIN |
 |
E.Coli Dsba In Complex With 4-Phenyl-2-(3-Phenylpropyl)Thiazole-5-Carboxylic Acid
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-04-12 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: QVP, CU |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN Ligands: CA, ZN |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: CU, 010 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: IMD |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: BYZ, PGE |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: PKN, CU |
 |
Crystal Structure Of Ecdsba In A Complex With (1-Methyl-1H-Pyrazol-5-Yl)Methanamine
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q2I, CU |

