Search Count: 28
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-02-22 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: REPLICATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: REPLICATION |
 |
Crystal Structure Of The Oxyr Regulatory Domain Of Shewanella Oneidensis Mr-1, Reduced Form
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-02-09 Classification: DNA BINDING PROTEIN |
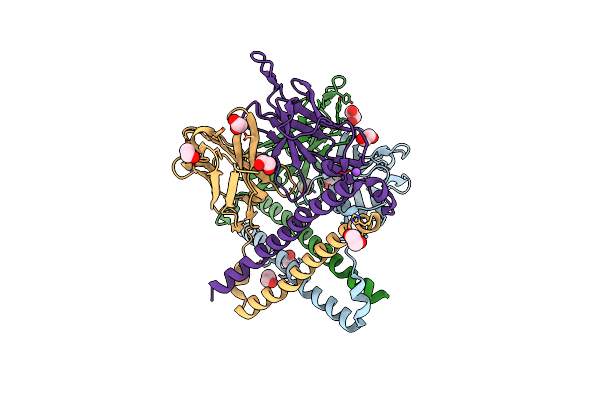 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-05-12 Classification: IMMUNE SYSTEM Ligands: EDO, CL, NA |
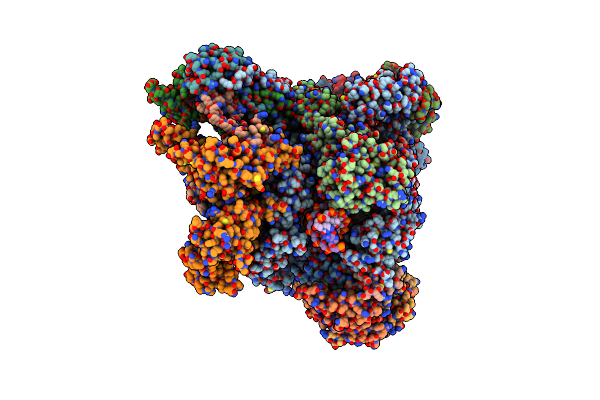 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-02-17 Classification: TRANSCRIPTION Ligands: ZN, SF4 |
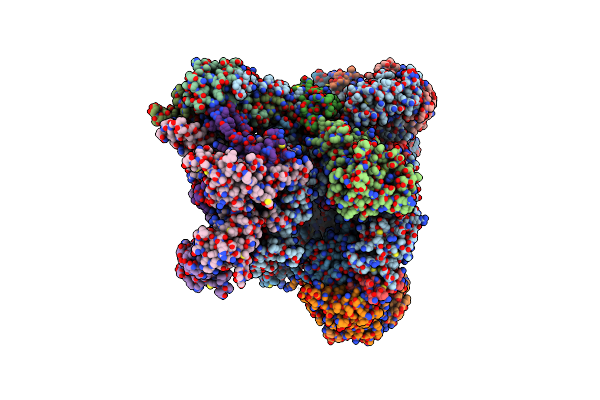 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-02-17 Classification: TRANSCRIPTION Ligands: ZN, SF4 |
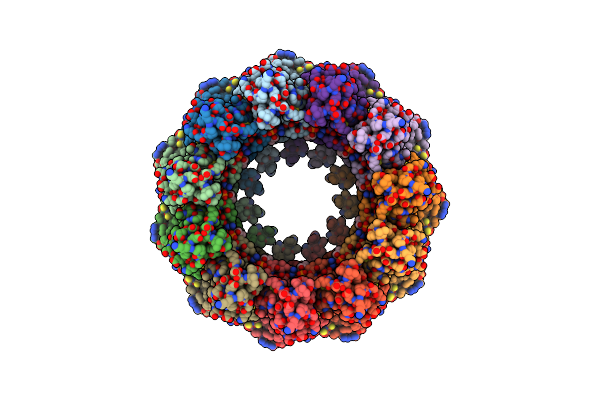 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-23 Classification: TRANSPORT PROTEIN Ligands: PA8 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-23 Classification: TRANSPORT PROTEIN Ligands: PA8 |
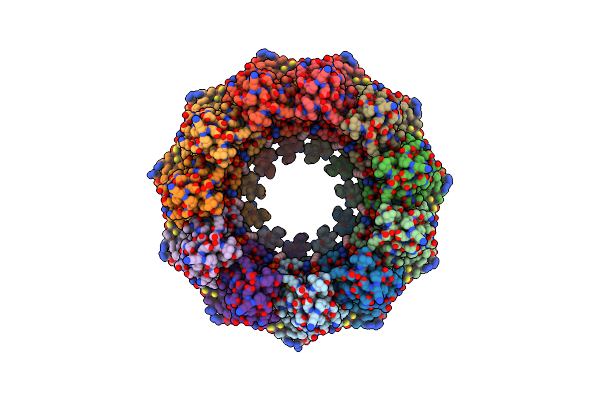 |
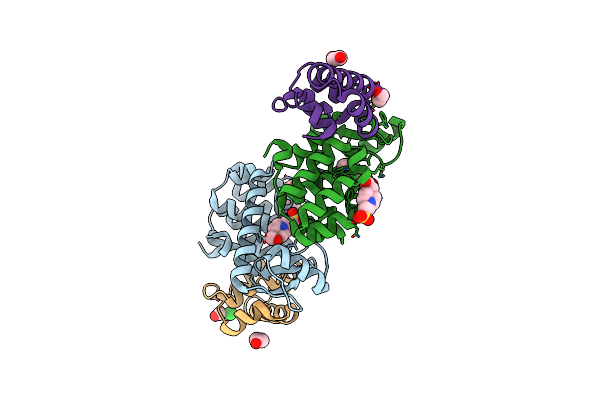
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-23 Classification: TRANSPORT PROTEIN Ligands: PA8 |
 |
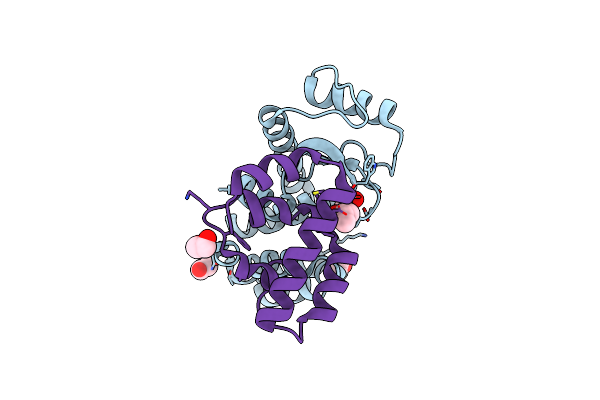
Crystal Structure Of Bacteriophage T4 Spackle And Lysozyme In Monoclinic Form
Organism: Escherichia virus t4
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: EDO, EPE, SIN, CL, FMT |
 |
Crystal Structure Of Bacteriophage T4 Spackle And Lysozyme In Orthorhombic Form
Organism: Escherichia virus t4
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: EDO, IPA |
 |
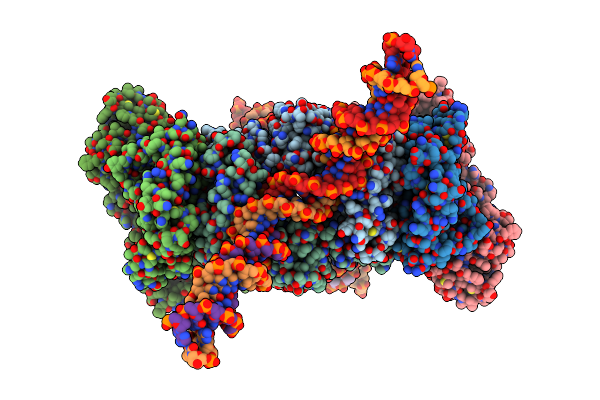
Crystal Structure Of T4 Protein Spackle As Determined By Native Sad Phasing
Organism: Escherichia virus t4
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2020-09-16 Classification: VIRAL PROTEIN Ligands: CL |
 |
Organism: Saccharolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2), Human t-cell leukemia virus type i, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, MG |
 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440), Methanocaldococcus jannaschii
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440), Escherichia coli, Methanocaldococcus jannaschii
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-09-26 Classification: LIGASE Ligands: GOL |
 |
Crystal Structure Of The Processivity Clamp Gp45 Complexed With Recognition Peptide Of Ligase From Bacteriophage T4
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2018-09-26 Classification: GENE REGULATION Ligands: EDO |
 |
Crystal Structure Of The Ligase From Bacteriophage T4 Complexed With Dna Intermediate
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-09-19 Classification: LIGASE/DNA Ligands: GOL, 1PE, DTT, CL, MG, AMP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2018-09-05 Classification: SIGNALING PROTEIN/inhibitor Ligands: HRA, IMD, EDO |

