Search Count: 47
 |
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: CHAPERONE |
 |
 |
 |
 |
The Structure Of Htpg M Domain In Complex With Unstructured D131D Binding Site B
Organism: Escherichia coli, Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: CHAPERONE |
 |
The Structure Of Htpg M Domain In Complex With Unstructured D131D Binding Site A
Organism: Escherichia coli, Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: CHAPERONE |
 |
Organism: Escherichia coli, Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: CHAPERONE |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: RNA |
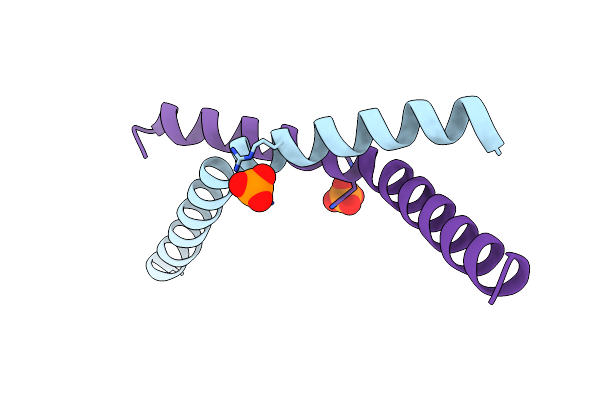 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-01-18 Classification: CHAPERONE Ligands: PO4 |
 |
Organism: Yersinia pseudotuberculosis serotype i (strain ip32953)
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2022-12-21 Classification: TOXIN |
 |
Structure Of Candida Albicans Fructose-1,6-Bisphosphate Aldolase Complexed With G3P
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-02-23 Classification: LYASE Ligands: ZN, G3H |
 |
Structure Of Candida Albicans Fructose-1,6-Bisphosphate Aldolase Mutation C157S With Cn39
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2022-02-23 Classification: LYASE Ligands: ZN, EDO, 6Y3 |
 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2020-12-30 Classification: LYASE Ligands: ZN, EDO |
 |
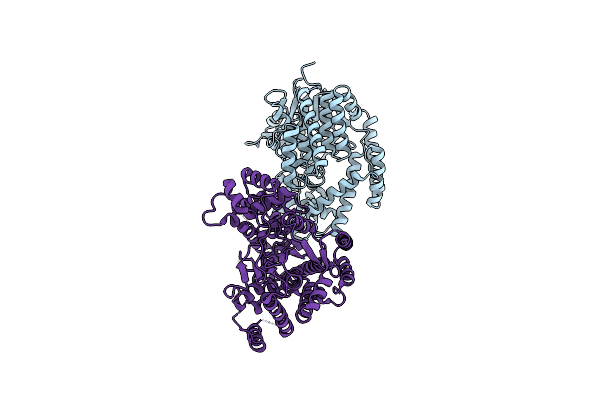
Crystal Structure Of The Udp-Bound Glycosyltransferase Domain From The Ygt Toxin
Organism: Yersinia mollaretii (strain atcc 43969 / dsm 18520 / cip 103324 / cny 7263 / waip 204)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-03-18 Classification: TOXIN Ligands: UDP, MN, K, EDO |
 |
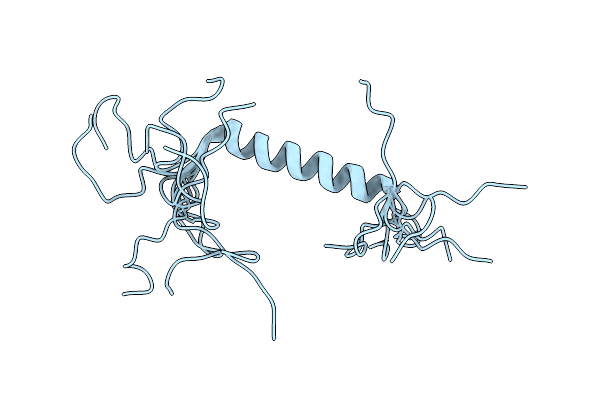
Crystal Structure Of The Ligand-Free Glycosyltransferase Domain From The Ygt Toxin
Organism: Yersinia mollaretii (strain atcc 43969 / dsm 18520 / cip 103324 / cny 7263 / waip 204)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-03-18 Classification: TOXIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-11-27 Classification: NUCLEAR PROTEIN |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-09-13 Classification: PROTEIN TRANSPORT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2017-09-13 Classification: PROTEIN TRANSPORT |
 |
Crystal Structure Of The Hepatitis C Virus Ns5B Rna-Dependent Rna Polymerase In Complex With 2-(4-Fluorophenyl)-N-Methyl-6-[(Methylsulfonyl)Amino]-5-(Propan-2-Yloxy)-1-Benzofuran-3-Carboxamide
Organism: Hepatitis c virus genotype 1b (isolate con1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-05-10 Classification: TRANSFERASE Ligands: 23E, 2N5, SO4, GOL, PG4 |

