Search Count: 41
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.81 Å Release Date: 2016-03-09 Classification: STRUCTURAL PROTEIN Ligands: DMS |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-01-14 Classification: TRANSFERASE/DNA Ligands: MG, 1FZ |
 |
Organism: Japanese encephalitis virus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Hypothetical Protein Pf0907 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-10 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Crystal Structure Of Hypothetical Protein Pf0907 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-12-10 Classification: UNKNOWN FUNCTION Ligands: CL, IMD |
 |
Crystal Structure Of Thermus Thermophilus Thioredoxin Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-12-10 Classification: ELECTRON TRANSPORT Ligands: CL |
 |
Hypothetical Protein Pf1117 From Pyrococcus Furiosus: Structure Solved By Sulfur-Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-12-10 Classification: ISOMERASE Ligands: GSH, LVJ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-12-10 Classification: CELL CYCLE |
 |
Crystal Structure Of The Exonuclease Domain Of Qip (Qde-2 Interacting Protein) Solved By Native-Sad Phasing.
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2014-12-10 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
Conserved Hypothetical Protein Pf1771 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-10 Classification: OXIDOREDUCTASE |
 |
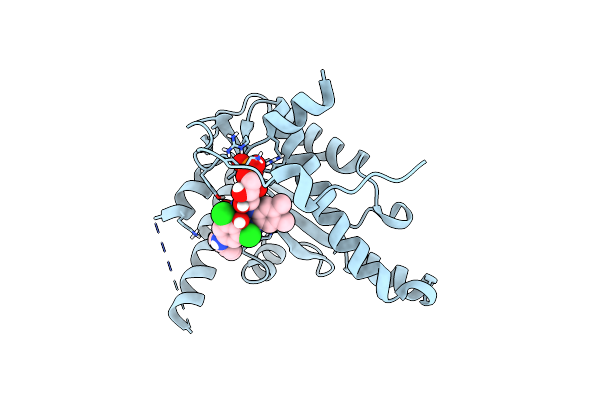
Nitroreductase Cind From Lactococcus Lactis In Complex With Chloramphenicol
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: FMN, CLM |
 |
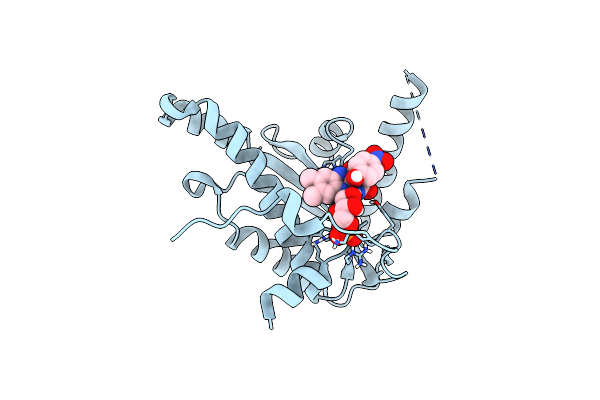
Nitroreductase Cind From Lactococcus Lactis In Complex With 2,6- Dichlorophenolindophenol
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: FMN, RLM |
 |
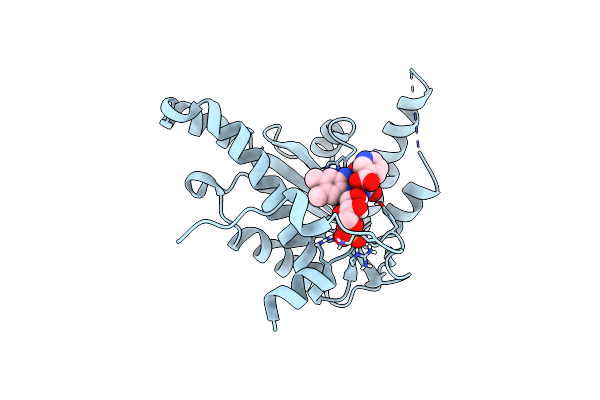
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: FMN, NPO |
 |
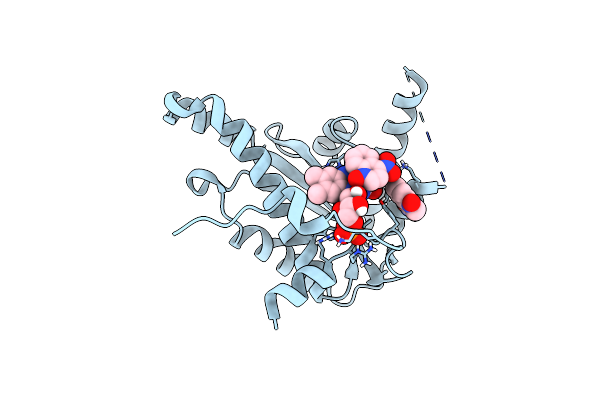
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: NIO, FMN |
 |
Nitroreductase Cind From Lactococcus Lactis In Complex With 4- Nitroquinoline 1-Oxide
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: FMN, YHX |
 |
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2014-01-22 Classification: VIRAL PROTEIN/ANTIVIRAL PROTEIN |
 |
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2013-12-11 Classification: MEMBRANE PROTEIN/INHIBITOR |
 |
Apo Form Of A Putative Sugar Kinase Mk0840 From Methanopyrus Kandleri (Monoclinic Space Group)
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2013-11-27 Classification: TRANSFERASE Ligands: K, CL, ACT |
 |
Apo Form Of A Putative Sugar Kinase Mk0840 From Methanopyrus Kandleri (Orthorhombic Space Group)
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-11-27 Classification: TRANSFERASE Ligands: K, CL, ACT |

