Search Count: 15
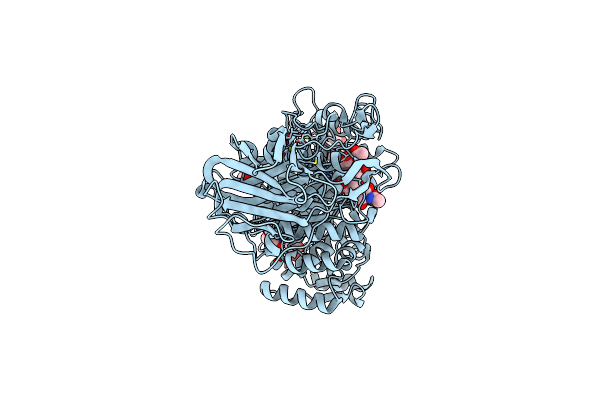 |
Ruminococcus Gnavus Atc29149 Endo-Beta-1,4-Galactosidase (Rggh98) E411A In Complex With Blood Group A (Bga Ii) Tetrasaccharide
Organism: Ruminococcus gnavus (strain atcc 29149 / vpi c7-9)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: GOL, TRS, CA, MG |
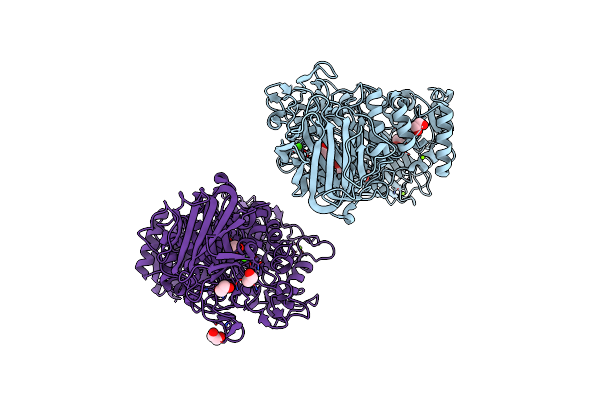 |
Organism: Ruminococcus gnavus (strain atcc 29149 / vpi c7-9)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: EDO, PEG, MG, CA |
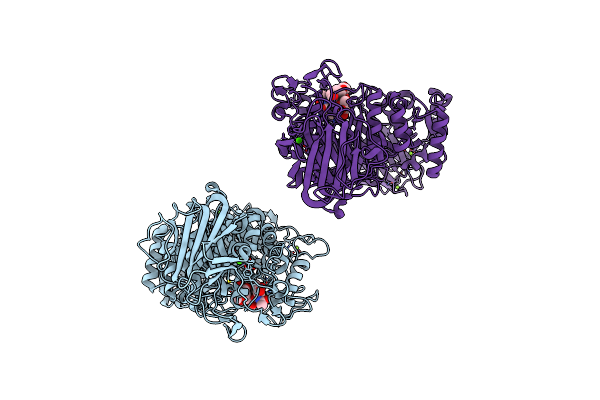 |
Ruminococcus Gnavus Atc29149 Endo-Beta-1,4-Galactosidase (Rggh98) In Complex With Blood Group A Trisaccharide
Organism: Ruminococcus gnavus (strain atcc 29149 / vpi c7-9)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: CA, MG, PGE |
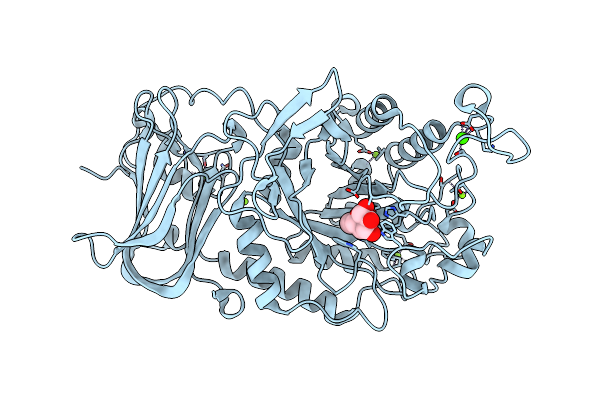 |
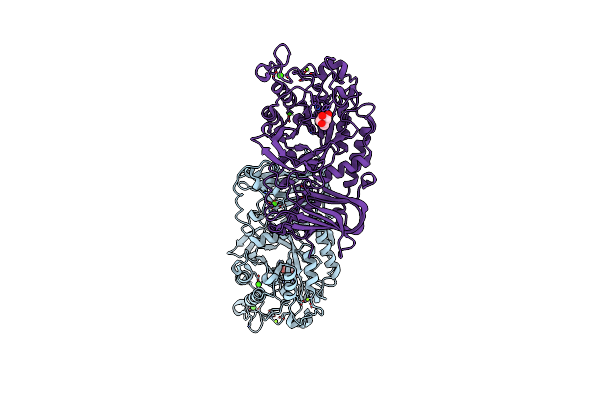
Organism: [ruminococcus] gnavus e1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: FUL, MG, CA |
 |
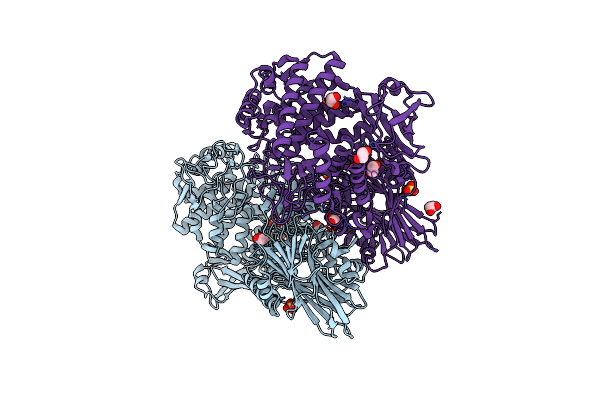
Ruminococcus Gnavus Gh29 Fucosidase E1_10125 D221A Mutant In Complex With Fucose
Organism: [ruminococcus] gnavus e1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: FUL, FUC, MG, CA, CL |
 |
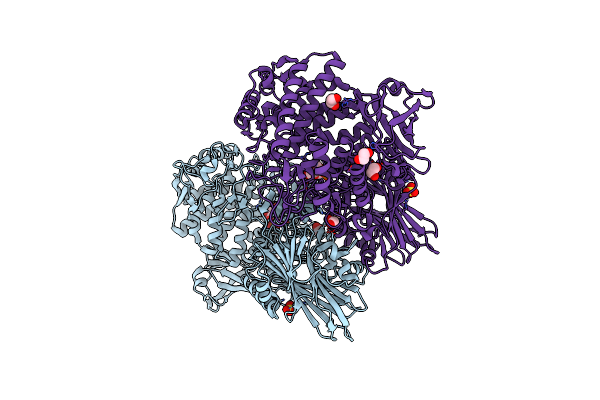
Organism: Paenibacillus sp. ym1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-06-13 Classification: HYDROLASE Ligands: SO4, EDO, CL |
 |
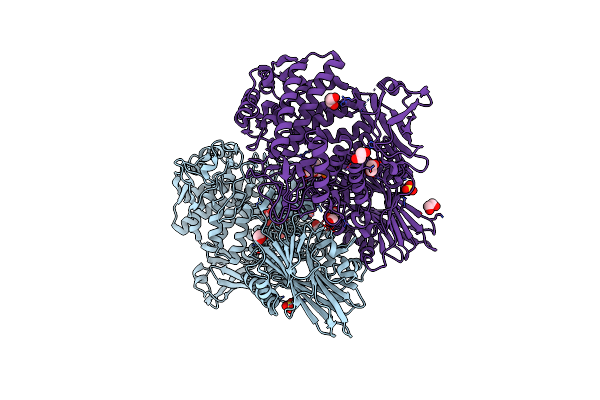
Paenibacillus Sp. Ym1 Laminaribiose Phosphorylase With Alpha-Glc-1-Phosphate Bound
Organism: Paenibacillus sp. ym1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-06-13 Classification: HYDROLASE Ligands: SO4, G1P, EDO, CL |
 |
Paenibacillus Sp. Ym1 Laminaribiose Phosphorylase With Alpha-Man-1-Phosphate Bound
Organism: Paenibacillus sp. ym1
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2018-06-13 Classification: HYDROLASE Ligands: SO4, M1P, EDO, CL |
 |
Organism: Lactobacillus reuteri atcc 53608
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-03-21 Classification: CELL ADHESION |
 |
Organism: Lactobacillus reuteri 100-23
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-03-21 Classification: CELL ADHESION |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-12-27 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-12-27 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2017-12-27 Classification: TRANSFERASE Ligands: EDO, MN, UDP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.64 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: UDP, MN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: UD1, MN |

