Search Count: 25
 |
Crystal Structure Of A Mycobacteriophage Cluster A2 Immunity Repressor:Dna Complex
Organism: Mycobacterium phage tipsythetrex
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2022-07-20 Classification: GENE REGULATION/DNA |
 |
Crystal Structure Of A Mycobacteriophage Cluster A2 Immunity Repressor:Dna Complex
Organism: Mycobacterium phage tipsythetrex
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2022-07-20 Classification: GENE REGULATION/DNA |
 |
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-12-08 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-12-08 Classification: HYDROLASE Ligands: GD3 |
 |
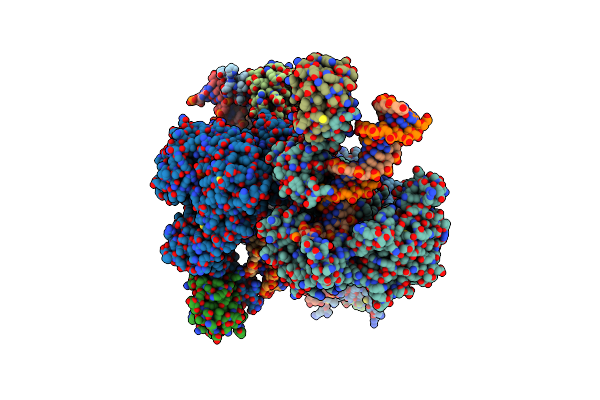
A Structural Characterization Of Poly(Aspartic Acid) Hydrolase-1 From Sphingomonas Sp. Kt-1.
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-12-09 Classification: HYDROLASE |
 |
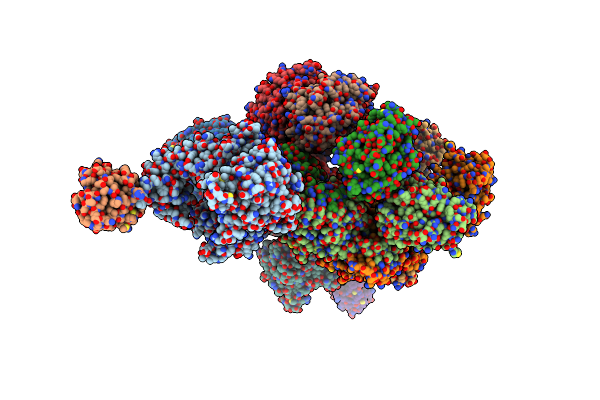
Structure Of T7 Dna Polymerase Bound To A Primer/Template Dna And A Peptide That Mimics The C-Terminal Tail Of The Primase-Helicase
Organism: Enterobacteria phage t7, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-03-04 Classification: TRANSFERASE/DNA BINDING PROTEIN/DNA Ligands: TTP, MG |
 |
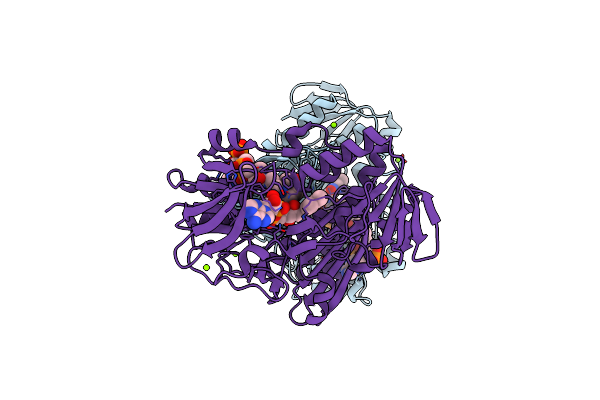
Organism: Enterobacteria phage t7, Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:4.80 Å Release Date: 2016-12-07 Classification: TRANSFERASE |
 |
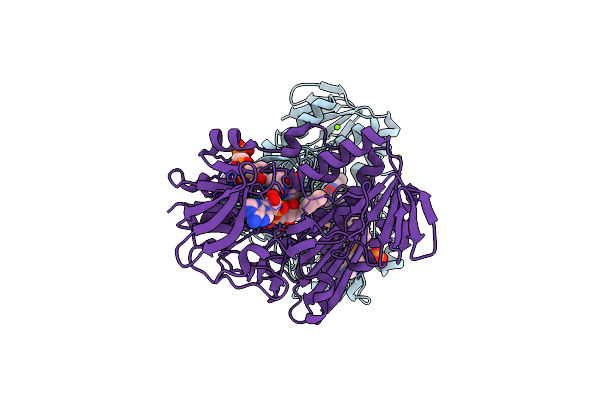
Crystal Structure Of Staphylococcus Aureus Bound With The Covalent Inhibitor Mevs-Coa
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, MG, CL, CA6 |
 |
Crystal Structure Of Staphylococcus Aureus Bound With The Covalent Inhibitor Pethyl-Vs-Coa
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, MG, CL, CA8 |
 |
Crystal Structure Of Staphylococcus Aureus Bound With The Covalent Inhibitor Etvc-Coa
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, MG, CAJ, CL |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE Ligands: MG, CL, COA, FAD |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE Ligands: MG, CL, COA, FAD |
 |
Crystal Structure Of The Y361F, Y419F Mutant Of Staphylococcus Aureus Coadr
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE Ligands: MG, CL, COA, FAD |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE Ligands: FAD, MG, CL |
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-11-17 Classification: OXIDOREDUCTASE Ligands: FAD, COA |
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2009-11-17 Classification: OXIDOREDUCTASE Ligands: FAD, COA, ADP |
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-11-17 Classification: OXIDOREDUCTASE Ligands: FAD, COA, ADP |
 |
Pyridine Nucleotide Complexes With Bacillus Anthracis Coenzyme A-Disulfide Reductase: A Structural Analysis Of Dual Nad(P)H Specificity
Organism: Bacillus anthracis str.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-22 Classification: OXIDOREDUCTASE Ligands: COA, FAD |
 |
Pyridine Nucleotide Complexes With Bacillus Anthracis Coenzyme A-Disulfide Reductase: A Structural Analysis Of Dual Nad(P)H Specificity
Organism: Bacillus anthracis str.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-04-22 Classification: OXIDOREDUCTASE Ligands: COA, FAD |
 |
Pyridine Nucleotide Complexes With Bacillus Anthracis Coenzyme A-Disulfide Reductase: A Structural Analysis Of Dual Nad(P)H Specificity
Organism: Bacillus anthracis str.
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2008-04-22 Classification: OXIDOREDUCTASE Ligands: COA, FAD, NAD |

