Search Count: 48
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: GOL, PEG, NA |
 |
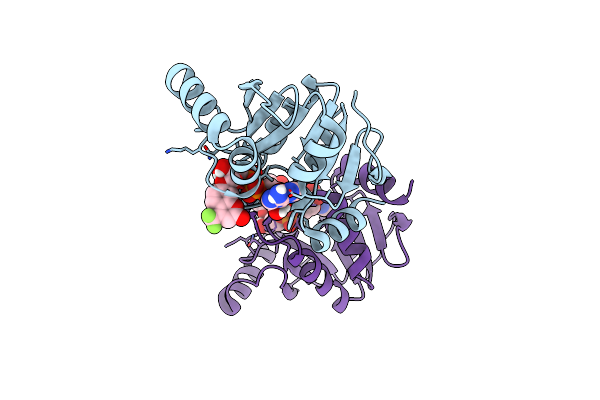
Crystal Structure Of Sars-Cov-2 (Covid-19) Nsp3 Macrodomain In Complex With Ndpr
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: A1AH3 |
 |
Crystal Structure Of Sars-Cov-2 (Covid-19) Nsp3 Macrodomain In Complex With Tfmu-Adpr
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-05-17 Classification: VIRAL PROTEIN Ligands: ZJ3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-12-29 Classification: HYDROLASE, Lyase Ligands: SF4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-12-29 Classification: HYDROLASE, Lyase Ligands: SF4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-01-01 Classification: HYDROLASE, LYASE Ligands: TRS |
 |
Organism: Acanthamoeba polyphaga mimivirus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2014-01-01 Classification: HYDROLASE, LYASE/DNA |
 |
Organism: Acanthamoeba polyphaga mimivirus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2013-10-30 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: ZN, IOD |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-25 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-07-25 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-07-25 Classification: HYDROLASE/DNA |
 |
Crystal Structure Of Dna-Glycosylase Bound To Dna Containing 5-Hydroxyuracil
Organism: Acanthamoeba polyphaga mimivirus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-12-14 Classification: HYDROLASE/DNA Ligands: FMT |
 |
Crystal Structure Of Dna-Glycosylase Bound To Dna Containing Thymine Glycol
Organism: Acanthamoeba polyphaga mimivirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-12-14 Classification: HYDROLASE/DNA Ligands: GOL |
 |
Organism: Enterobacteria phage rb69
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-11-09 Classification: Transferase/DNA Ligands: SO4 |
 |
Organism: Enterobacteria phage rb69
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2011-11-09 Classification: Transferase/DNA Ligands: SO4 |
 |
Organism: Escherichia phage rb69, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-11-09 Classification: Transferase/DNA Ligands: SO4 |
 |
Organism: Escherichia phage rb69, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2011-11-09 Classification: Transferase/DNA Ligands: SO4 |
 |
Crystal Structure Of A Replicative Dna Polymerase Bound To Dna Containing Thymine Glycol
Organism: Enterobacteria phage rb69
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2011-08-10 Classification: TRANSFERASE/DNA |
 |
Crystal Structure Of A Replicative Dna Polymerase Bound To Dna Containing Thymine Glycol
Organism: Enterobacteria phage rb69
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2011-08-10 Classification: TRANSFERASE/DNA Ligands: SO4 |

