Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
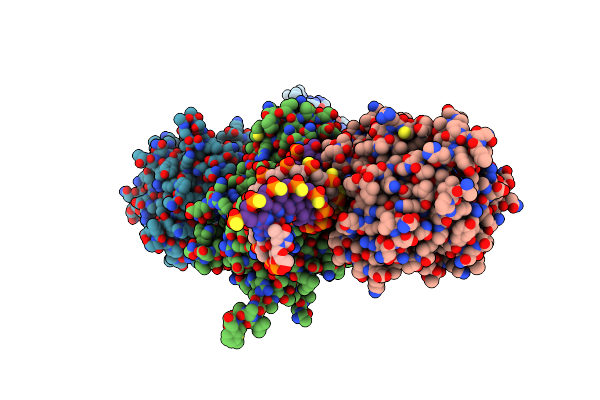 |
Crystal Structure Of Saccharomyces Cerevisiae Apn2 Catalytic Domain E59Q/D222N Mutant In Complex With Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2022-09-28 Classification: HYDROLASE/DNA Ligands: TAR, CA, TLA, CIT, EDO |
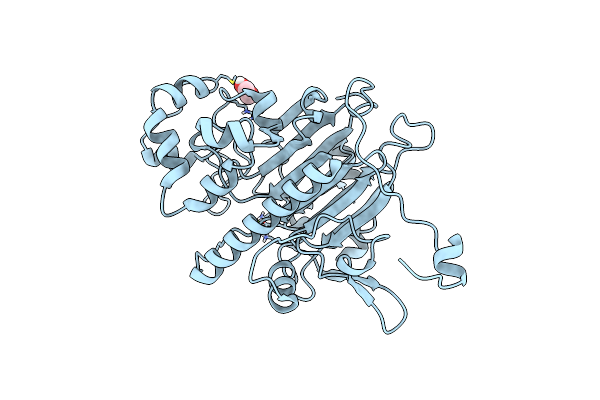 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: MG, CL, EDO |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-05-06 Classification: HYDROLASE Ligands: MG, MES, BU1, EDO |
 |
Crystal Structure Of Clostridium Perfringens Beta-Glucuronidase Bound With A Novel, Potent Inhibitor 4-(8-(Piperazin-1-Yl)-1,2,3,4-Tetrahydro-[1,2,3]Triazino[4',5':4,5]Thieno[2,3-C]Isoquinolin-5-Yl)Morpholine
Organism: Clostridium perfringens (strain 13 / type a), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-04-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: FJV |
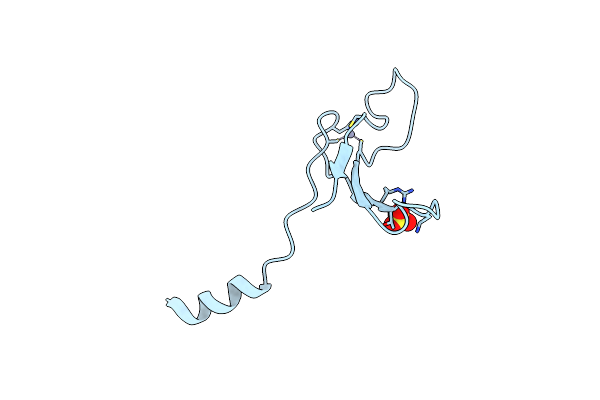 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-01-11 Classification: LYASE Ligands: ZN, SO4 |
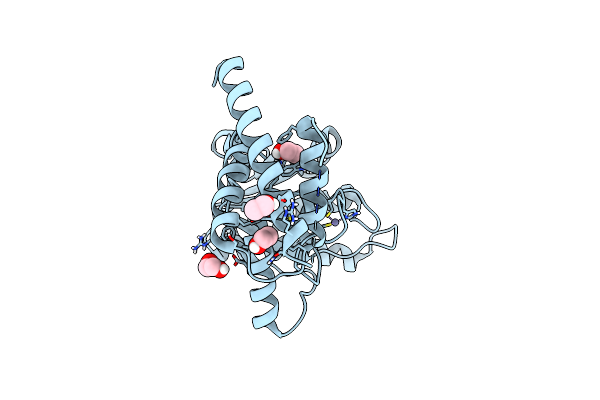 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-07-13 Classification: HYDROLASE Ligands: ZN, EDO, EOH |
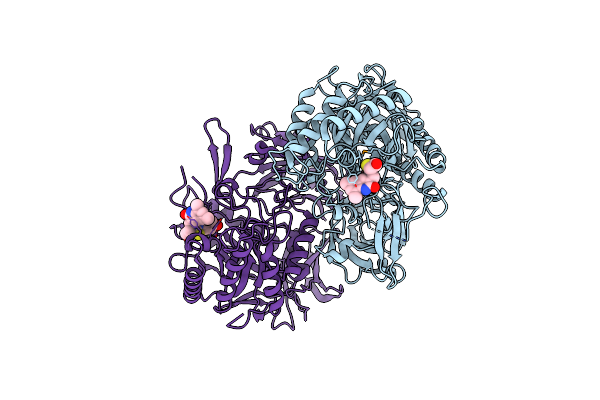 |
Structure Of E. Coli Beta-Glucuronidase Bound With A Novel, Potent Inhibitor 1-((6,8-Dimethyl-2-Oxo-1,2-Dihydroquinolin-3-Yl)Methyl)-1-(2-Hydroxyethyl)-3-(4-Hydroxyphenyl)Thiourea
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2015-10-14 Classification: Hydrolase/Hydrolase Inhibitor Ligands: 57Z |
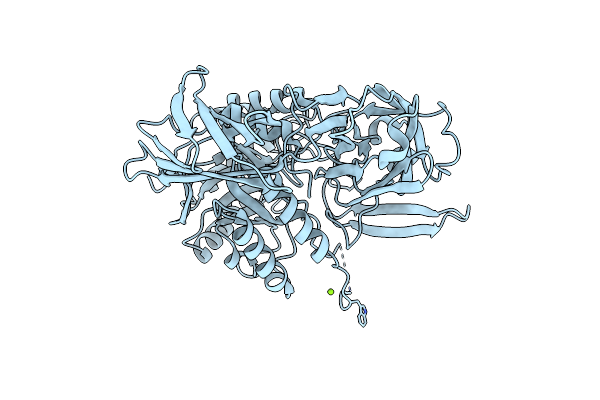 |
Crystal Structure Of Streptococcus Agalactiae Beta-Glucuronidase In Space Group I222
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: MG |
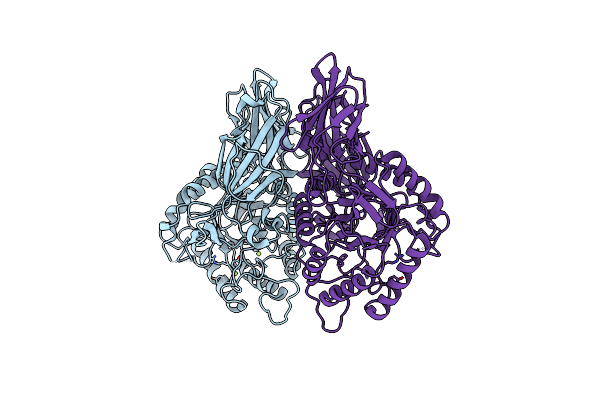 |
Crystal Structure Of Streptococcus Agalactiae Beta-Glucuronidase In Space Group P21212
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: MG |
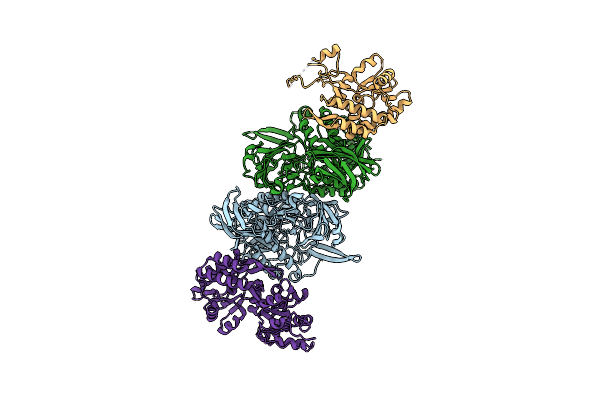 |
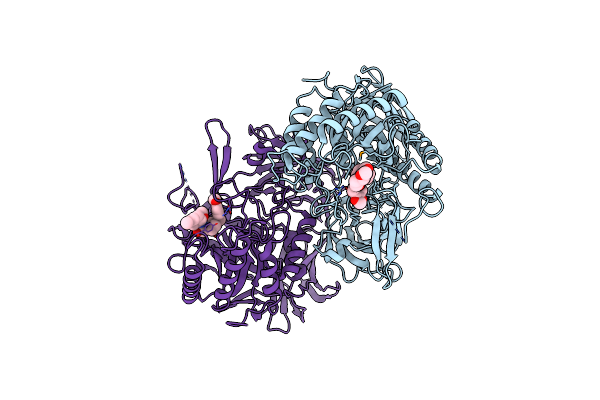
Organism: Clostridium perfringens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-09-17 Classification: HYDROLASE |
 |
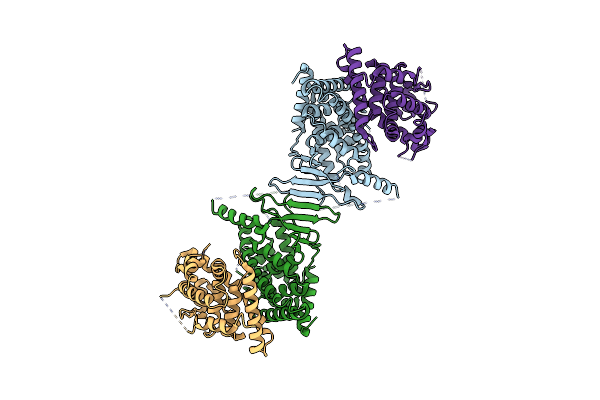
Structure Of E. Coli Beta-Glucuronidase Bound With A Novel, Potent Inhibitor 2-[4-(1,3-Benzodioxol-5-Ylmethyl)Piperazin-1-Yl]-N-[(1S,2S,5S)-2,5-Dimethoxycyclohexyl]Acetamide
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2013-08-28 Classification: hydrolase/hydrolase inhibitor Ligands: 1KV |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-08-21 Classification: Retinoic acid-binding protein Ligands: MG |
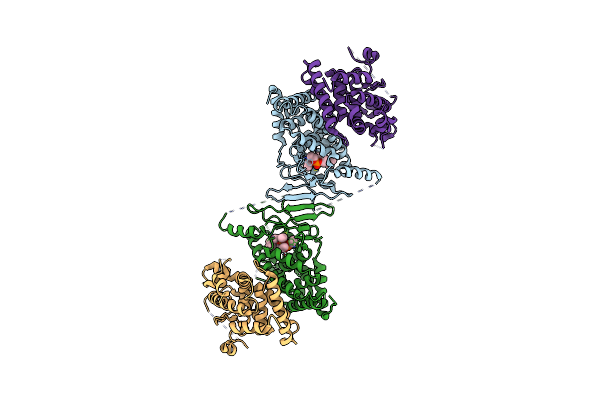 |
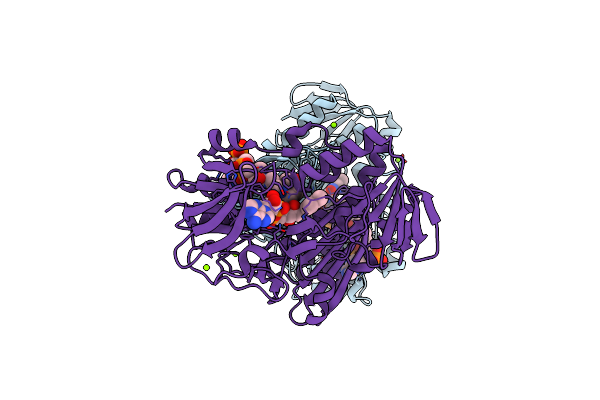
Crystal Structure Of The Sr12813-Bound Pxr/Rxralpha Lbd Heterotetramer Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-08-21 Classification: Retinoic acid-binding protein Ligands: SRL |
 |
Crystal Structure Of Staphylococcus Aureus Bound With The Covalent Inhibitor Mevs-Coa
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, MG, CL, CA6 |
 |
Crystal Structure Of Staphylococcus Aureus Bound With The Covalent Inhibitor Pethyl-Vs-Coa
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, MG, CL, CA8 |
 |
Crystal Structure Of Staphylococcus Aureus Bound With The Covalent Inhibitor Etvc-Coa
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, MG, CAJ, CL |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE Ligands: MG, CL, COA, FAD |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE Ligands: MG, CL, COA, FAD |
 |
Crystal Structure Of The Y361F, Y419F Mutant Of Staphylococcus Aureus Coadr
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE Ligands: MG, CL, COA, FAD |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-17 Classification: OXIDOREDUCTASE Ligands: FAD, MG, CL |