Search Count: 13
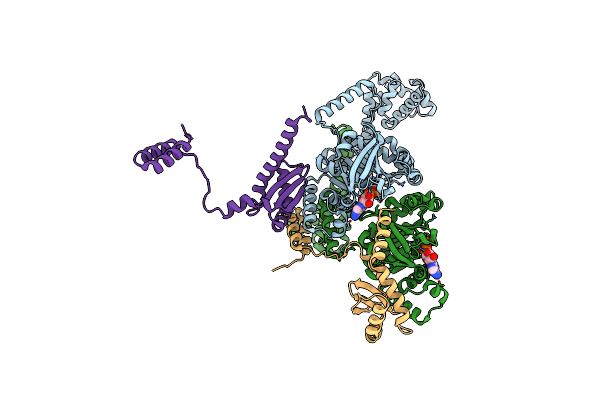 |
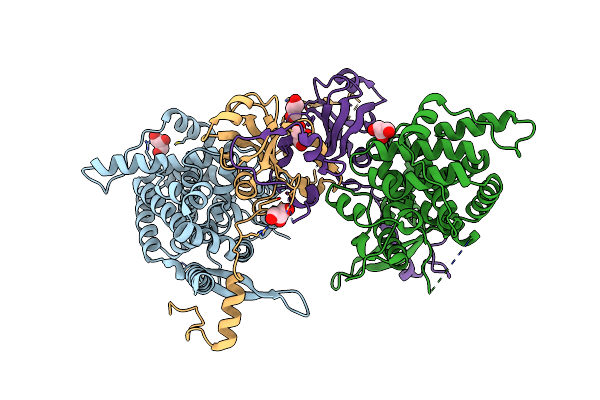
Crystal Structure Of The Vibrio Cholerae Replicative Helicase (Vcdnab) In Complex With Its Loader Protein (Vcdcia)
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-02-15 Classification: REPLICATION Ligands: ADP, MG |
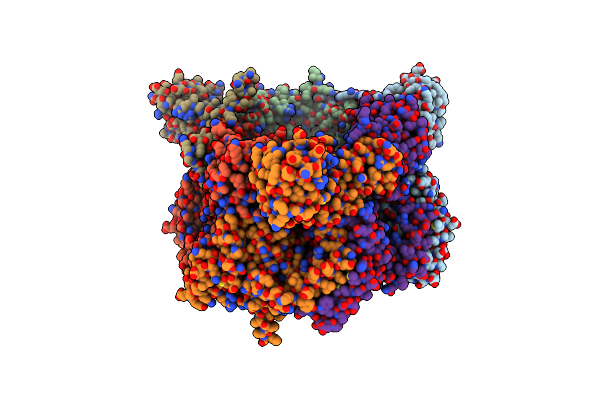 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2022-05-25 Classification: REPLICATION |
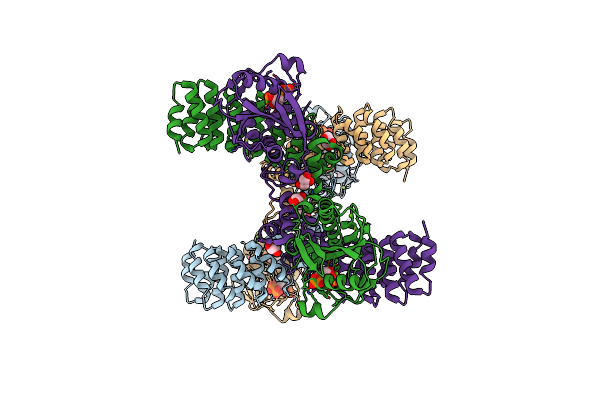 |
Crystal Structure Of Helicobacter Pylori Comf Fused To An Artificial Alpharep Crystallization Helper(Named B2)
Organism: Synthetic construct, Helicobacter pylori (strain atcc 700392 / 26695)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-04-06 Classification: RECOMBINATION Ligands: ZN, PRP, MG, GOL |
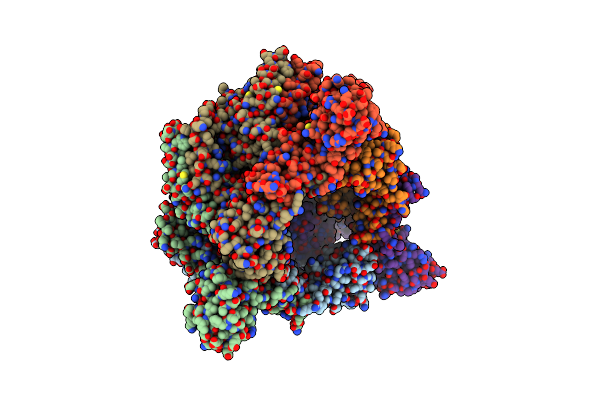 |
Crystal Structure Of The Vibrio Cholerae Replicative Helicase (Dnab) With Gdp-Alf4
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2021-04-28 Classification: REPLICATION Ligands: GDP, MG, ALF |
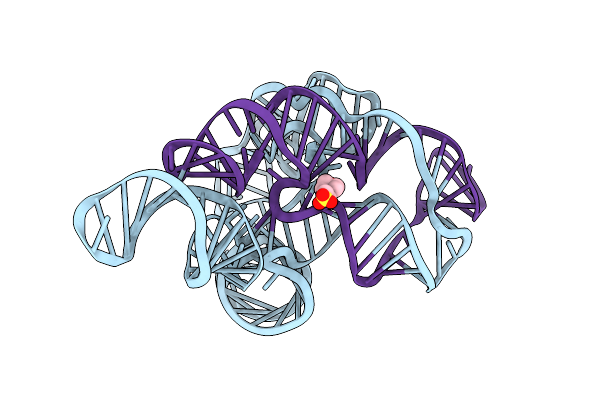 |
Organism: Didymium iridis
Method: X-RAY DIFFRACTION Resolution:3.34 Å Release Date: 2018-12-05 Classification: RNA Ligands: MG, MES |
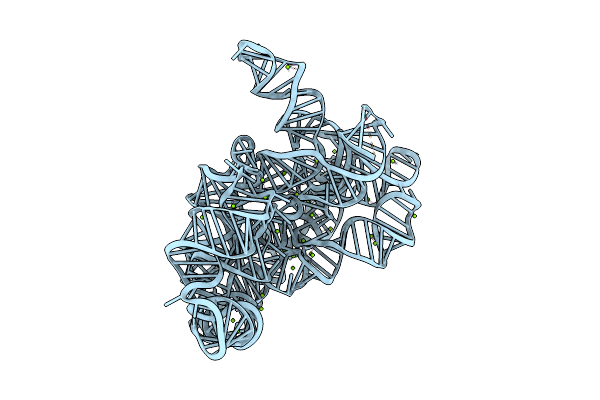 |
Structure Of The Lariat Form Of A Chimeric Derivative Of The Oceanobacillus Iheyensis Group Ii Intron In The Presence Of Nh4+ And Mg2+.
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2016-12-14 Classification: RNA Ligands: MG, NH4 |
 |
Structure Of The Lariat Form Of A Chimeric Derivative Of The Oceanobacillus Iheyensis Group Ii Intron In The Presence Of Nh4+, Mg2+ And An Inactive 5' Exon.
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2016-12-14 Classification: RNA Ligands: MG, NH4 |
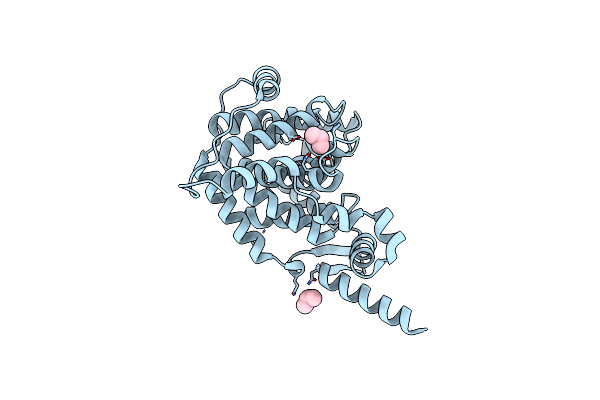 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-11-30 Classification: CELL CYCLE Ligands: IPA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-11-30 Classification: CELL CYCLE Ligands: GOL |
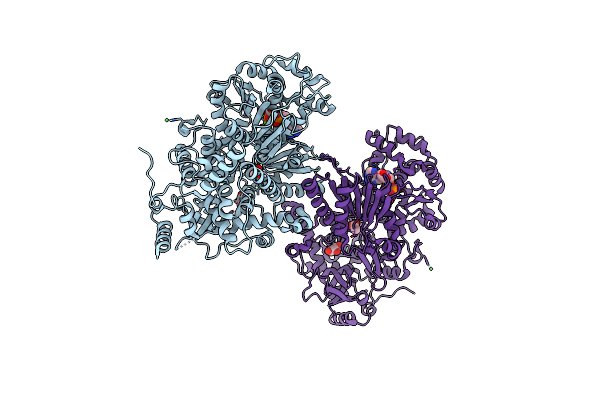 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-06-09 Classification: HYDROLASE Ligands: ADP, MG, NI, ACT, GOL |
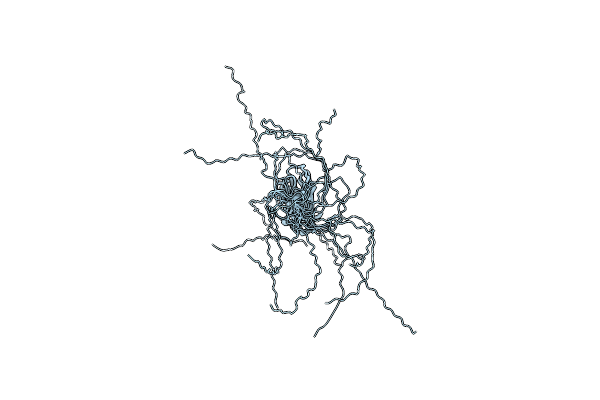 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2009-05-05 Classification: STRUCTURAL PROTEIN |
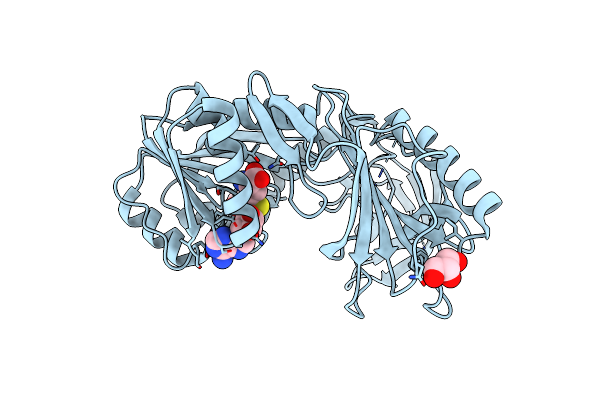 |
The Crystal Structure Of Pyrococcus Abyssi Trna (Uracil-54, C5)- Methyltransferase In Complex With S-Adenosyl-L-Homocysteine
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-08-05 Classification: TRANSFERASE Ligands: SAH, GOL |
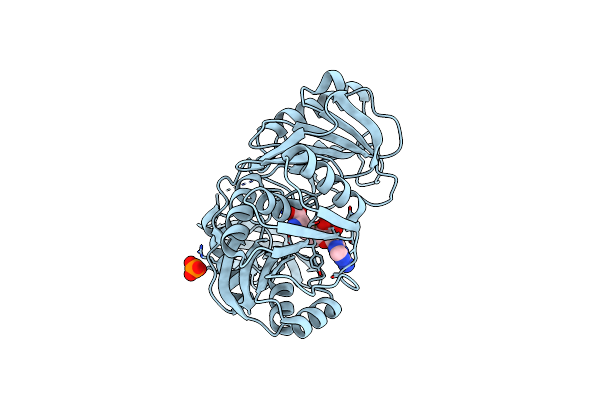 |
The Crystal Structure Of Pyrococcus Abyssi Trna (Uracil-54, C5)- Methyltransferase In Complex With S-Adenosyl-L-Homocysteine
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-08-05 Classification: TRANSFERASE Ligands: SAH, PO4 |

