Search Count: 145
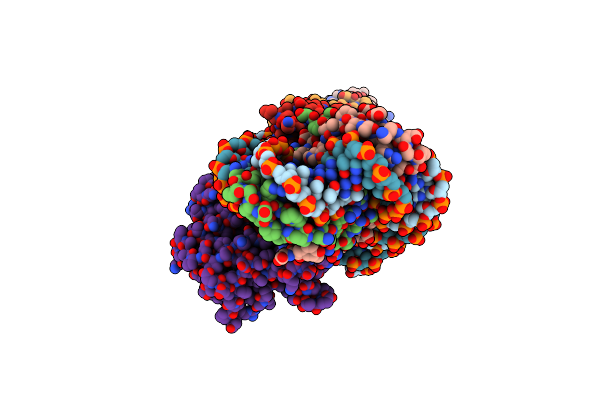 |
Cryoem Structure Of The F Plasmid Relaxosome In Its Pre-Initiation State, Derived From The Ds-27_+143-R Locally-Refined Map 3.76 A
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN |
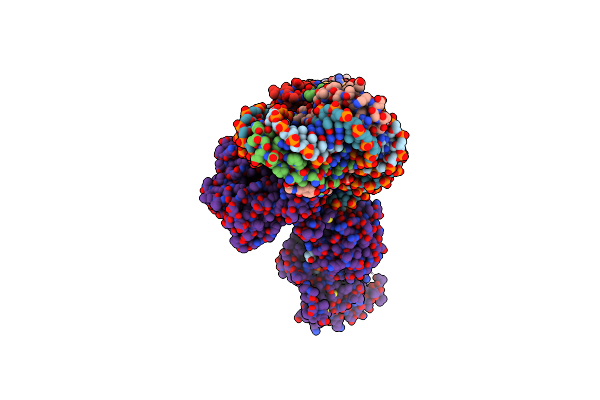 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R Locally-Refined 3.45 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
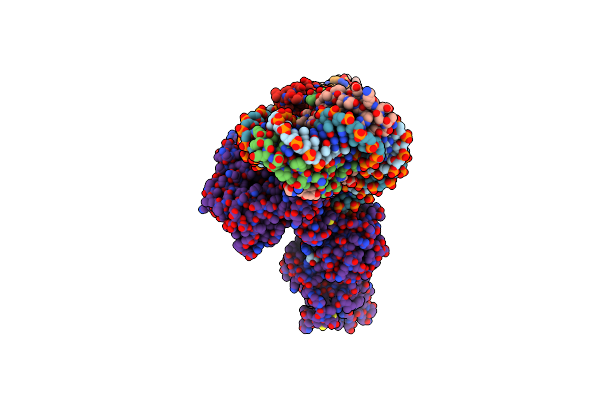 |
Cryoem Structure Of The F Plasmid Relaxosome With Truncated Trai1-863 In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R_Deltaah+Ctd Locally-Refined 3.42 A Map
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
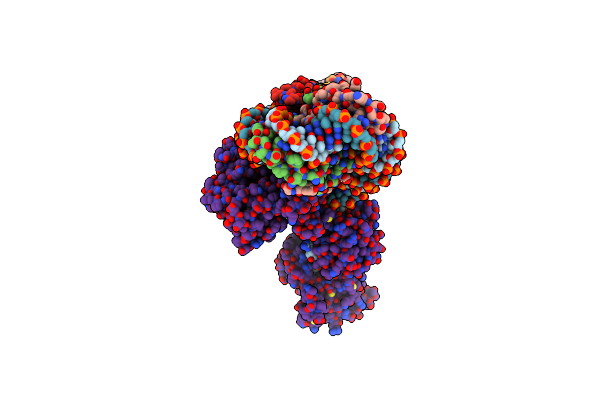 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Without Accessory Protein Tram. Derived From The Ss-27_+8Ds+9_+143-R_Deltatram Locally-Refined 2.94 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Orit Dna Ss-27_+3Ds+4_+143 And Trai Its Te Mode, Derived From Ss-27_+3Ds+4_+143-R Locally-Refined 3.68 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Orit Dna Ss-27_-3Ds-2_+143 And Trai Its Te Mode, Derived From Ss-27_-3Ds-2_+143-R Locally-Refined 3.42 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of The R388 Plasmid Conjugative Pilus Reveals A Helical Polymer Characterised By An Unusual Pilin/Phospholipid Binary Complex
Organism: Escherichia coli hb101
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: PROTEIN FIBRIL Ligands: LHG |
 |
O-Layer Structure (Trwh/Virb7, Trwf/Virb9Ctd, Trwe/Virb10Ctd) Of The Outer Membrane Core Complex From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
I-Layer Structure (Trwf/Virb9Ctd, Trwe/Virb10Ctd) Of The Outer Membrane Core Complex From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Conformation-A Of The Full-Length Outer Membrane Core Complex (Trwh/Virb7, Trwf/Virb9, Trwe/Virb10Ctd) From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
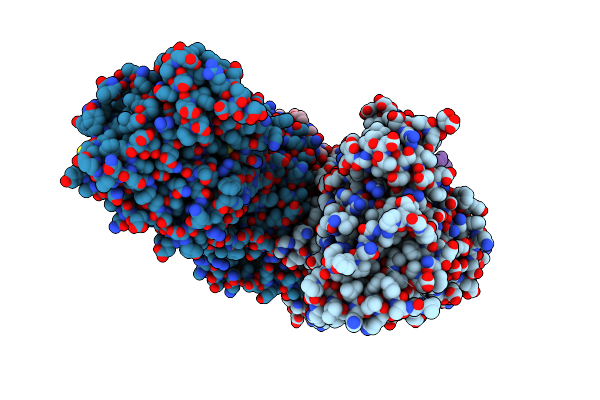
Conformation-B Of The Full-Length Outer Membrane Core Complex (Trwh/Virb7, Trwf/Virb9, Trwe/Virb10Ctd) From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
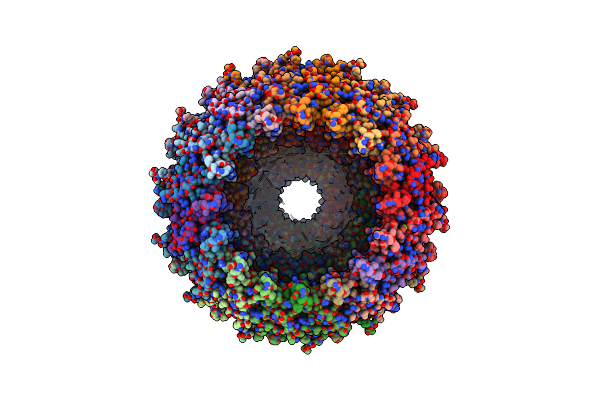 |
Conformation-C Of The Full-Length Outer Membrane Core Complex (Trwh/Virb7, Trwf/Virb9, Trwe/Virb10Ctd) From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
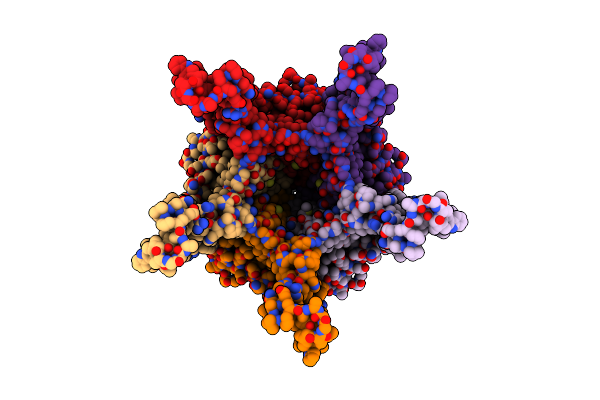 |
Stalk Complex Full-Length Structure (Trwj/Virb5-Trwi/Virb6) From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
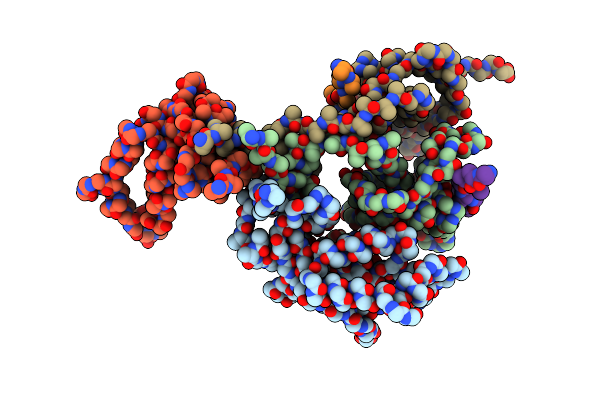 |
Arches-Protomer Complex Full-Length Structure (Trwj/Virb8) From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Extended Inner Membrane Complex (Imc) Protomer Structure (Trwm/Virb3-Trwk/Virb4-Trwi/Virb6-Trwg/Virb8-Trwe/Virb10) From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Stalk-Arches-Imc Structure From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: STRUCTURAL PROTEIN |
 |
2.5 A Cryo-Em Structure Of The In Vitro Fimd-Catalyzed Assembly Of Type 1 Pilus Rod
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: STRUCTURAL PROTEIN |
 |
O-Layer Structure (Trwh/Virb7, Trwf/Virb9Ctd, Trwe/Virb10Ctd) Of The Outer Membrane Core Complex From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
 |
I-Layer Structure (Trwf/Virb9Ntd, Trwe/Virb10Ntd) Of The Outer Membrane Core Complex From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |

