Search Count: 69
 |
The Structure Of The Glycopeptidase Catalytic Domain Including The Linker Of Amuc_1438
Organism: Akkermansia muciniphila
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: ZN, NA, EDO |
 |
Organism: Akkermansia muciniphila
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: ZN, CA, NA |
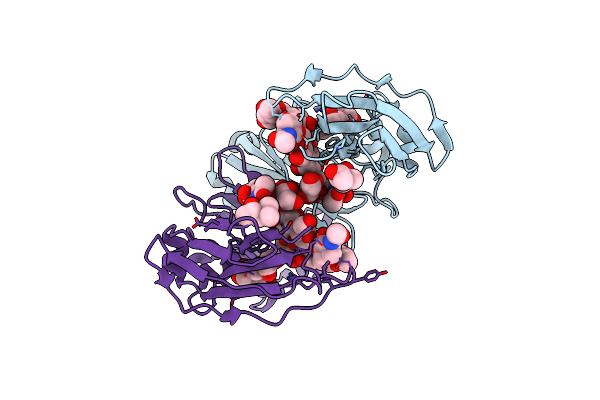 |
Crystal Structure Of A Human Fucosylated Igg1 Fc Expressed In Tobacco Plants (Nicotiana Benthamiana)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-17 Classification: IMMUNE SYSTEM Ligands: MPD |
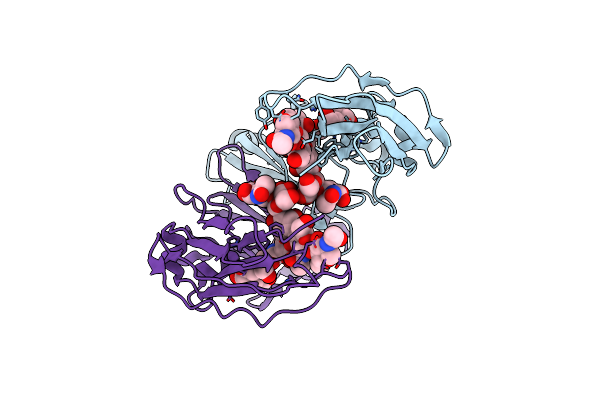 |
Crystal Structure Of A Human Afucosylated Igg1 Fc Expressed In Tobacco Plants (Nicotiana Benthamiana)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-02-17 Classification: IMMUNE SYSTEM |
 |
The Structure Of The Cbm32-1, Cbm32-2, And M60 Catalytic Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: ZN |
 |
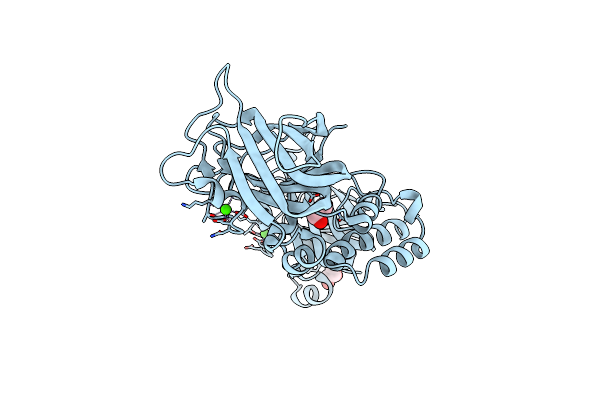
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb In Complex With Galnac
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, A2G, GOL |
 |
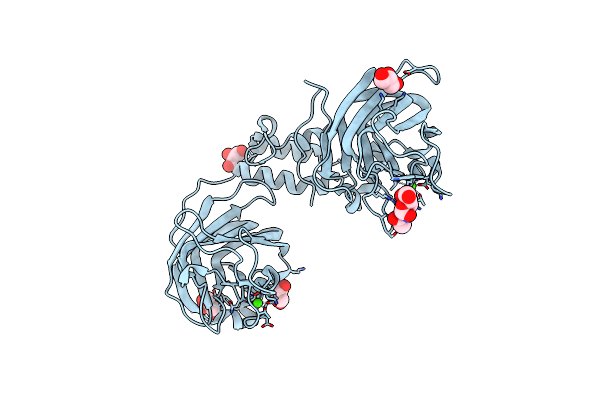
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL |
 |
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb In Complex With Galnac
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: A2G, CA, GOL |
 |
The Structure Of Cbm51-2 In Complex With Glcnac And Int Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, NAG, EDO, PO4, TRS |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
The Structure Of The M60 Catalytic Domain With The Cbm51-1 And Cbm51-2 Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2021-02-03 Classification: HYDROLASE |
 |
Organism: Mannheimia haemolytica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-02 Classification: MEMBRANE PROTEIN Ligands: SO4 |
 |
Organism: Mannheimia haemolytica
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-08-02 Classification: MEMBRANE PROTEIN Ligands: BR |
 |
Organism: Mannheimia haemolytica
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-08-02 Classification: MEMBRANE PROTEIN Ligands: SO4, CDP |
 |
Structure Of A Bacterial Polysialyltransferase In Complex With Fondaparinux
Organism: Mannheimia haemolytica
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2017-08-02 Classification: MEMBRANE PROTEIN Ligands: SO4 |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, EDO |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, EDO, PO4 |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, NI, NA, SER, A2G |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GOL, NA, ZN |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: EDO, ZN |

