Search Count: 17
 |
Organism: Nostoc punctiforme nies-2108
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-11-23 Classification: OXIDOREDUCTASE |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-07-10 Classification: HYDROLASE Ligands: 5GP, MN |
 |
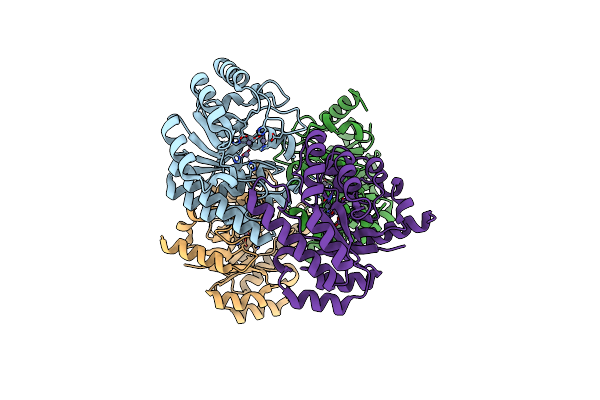
Crystal Structure Of Leucine Dehydrogenase From A Psychrophilic Bacterium Sporosarcina Psychrophila.
Organism: Sporosarcina psychrophila
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2013-02-06 Classification: OXIDOREDUCTASE |
 |
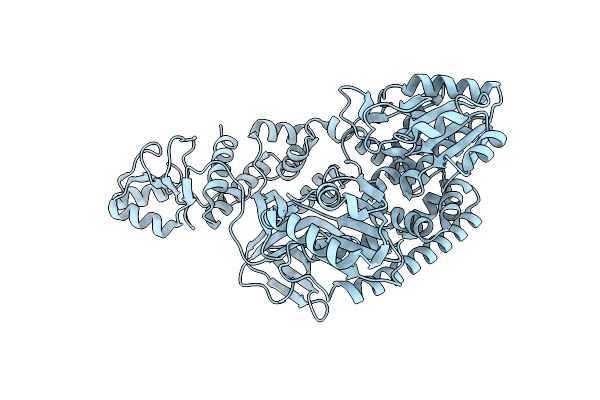
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-10-19 Classification: UNKNOWN FUNCTION Ligands: ZN |
 |
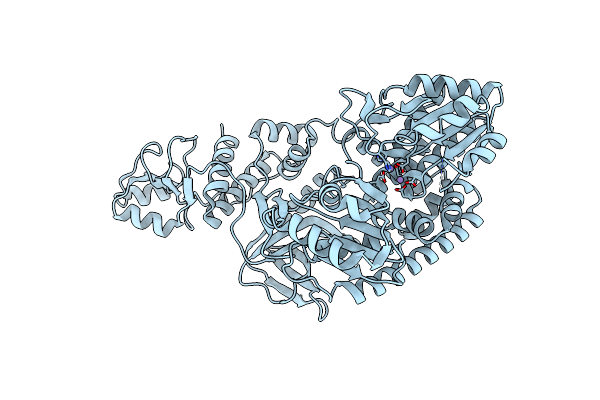
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-10-19 Classification: UNKNOWN FUNCTION Ligands: ZN |
 |
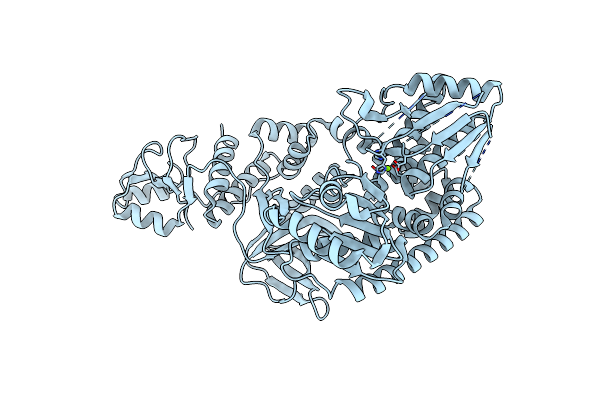
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-01-05 Classification: HYDROLASE |
 |
Crystal Structure Of Recj In Complex With Mn2+ From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-01-05 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Recj In Complex With Mg2+ From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2010-01-05 Classification: HYDROLASE Ligands: MG |
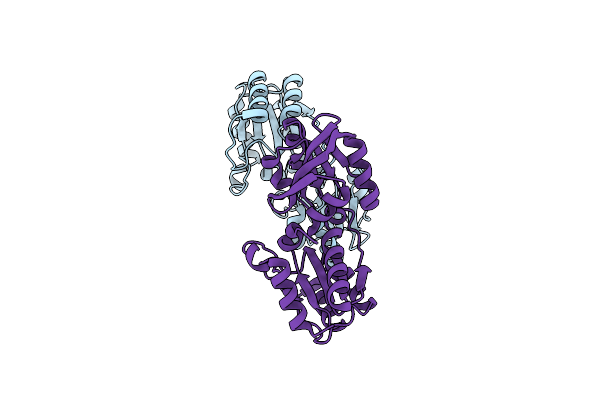 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-05-06 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
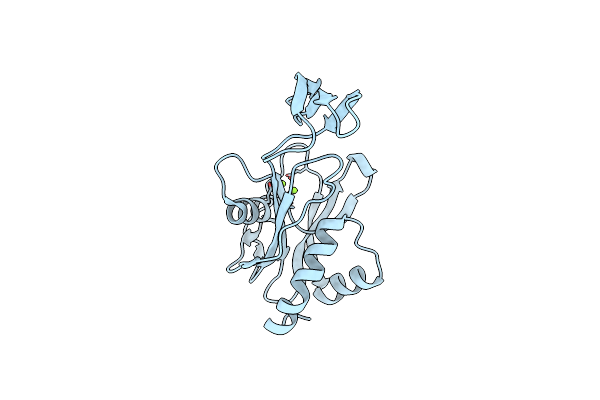 |
Crystal Structure Of Ndx2 In Complex With Mg2+ From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-02-26 Classification: HYDROLASE Ligands: MG |
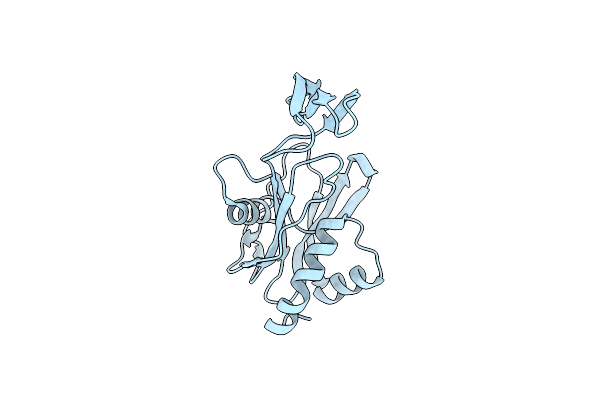 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2008-02-26 Classification: HYDROLASE |
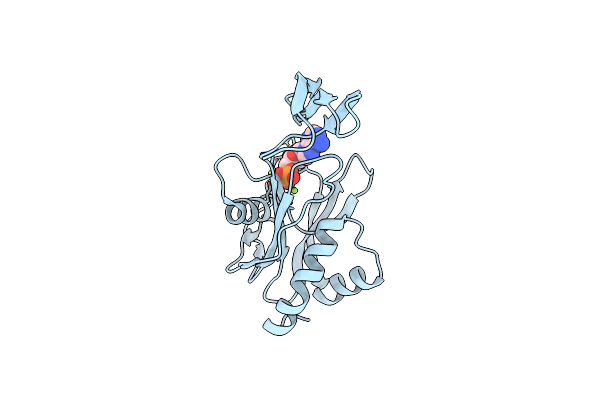 |
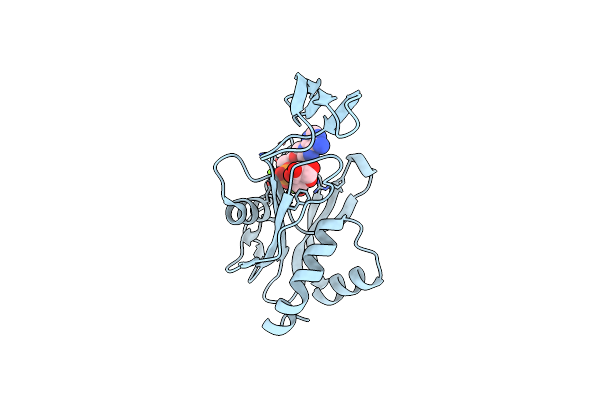
Crystal Structure Of Ndx2 In Complex With Mg2+ And Amp From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2008-02-26 Classification: HYDROLASE Ligands: MG, AMP |
 |
Crystal Structure Of Ndx2 In Complex With Mg2+ And Ampcpr From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2008-02-26 Classification: HYDROLASE Ligands: MG, RBY |
 |
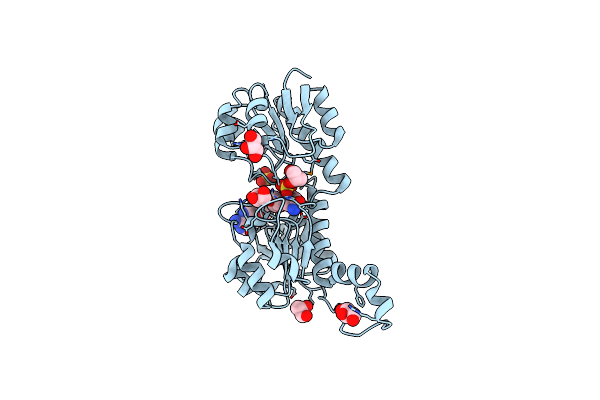
Crystal Structure Of The Hypothetical Sua5 Protein From Sulfolobus Tokodaii
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-01-15 Classification: RNA BINDING PROTEIN Ligands: MG, AMP |
 |
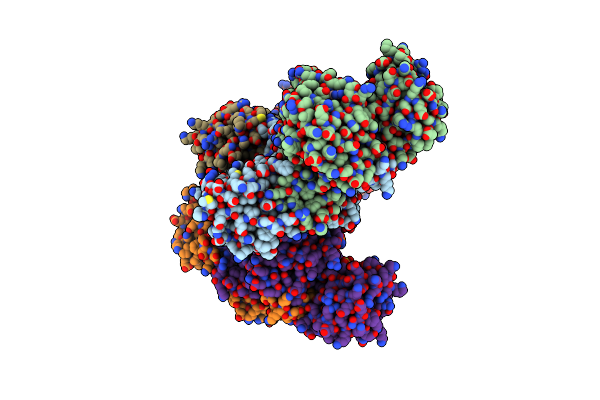
Crystal Structure Of Glyoxylate Reductase (Ph0597) From Pyrococcus Horikoshii Ot3, Complexed With Nadp (I41)
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-06-16 Classification: OXIDOREDUCTASE Ligands: SO4, NAP, GOL |
 |
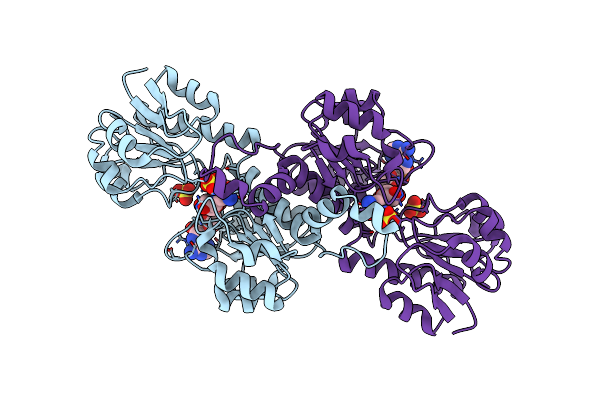
Crystal Structure Of Glyoxylate Reductase (Ph0597) From Pyrococcus Horikoshii Ot3, Complexed With Nadp (P1)
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2006-06-16 Classification: OXIDOREDUCTASE Ligands: SO4, NAP |
 |
Crystal Structure Of Glyoxylate Reductase (Ph0597) From Pyrococcus Horikoshii Ot3, Complexed With Nadp (P61)
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2006-06-16 Classification: OXIDOREDUCTASE Ligands: SO4, NAP |

