Search Count: 120
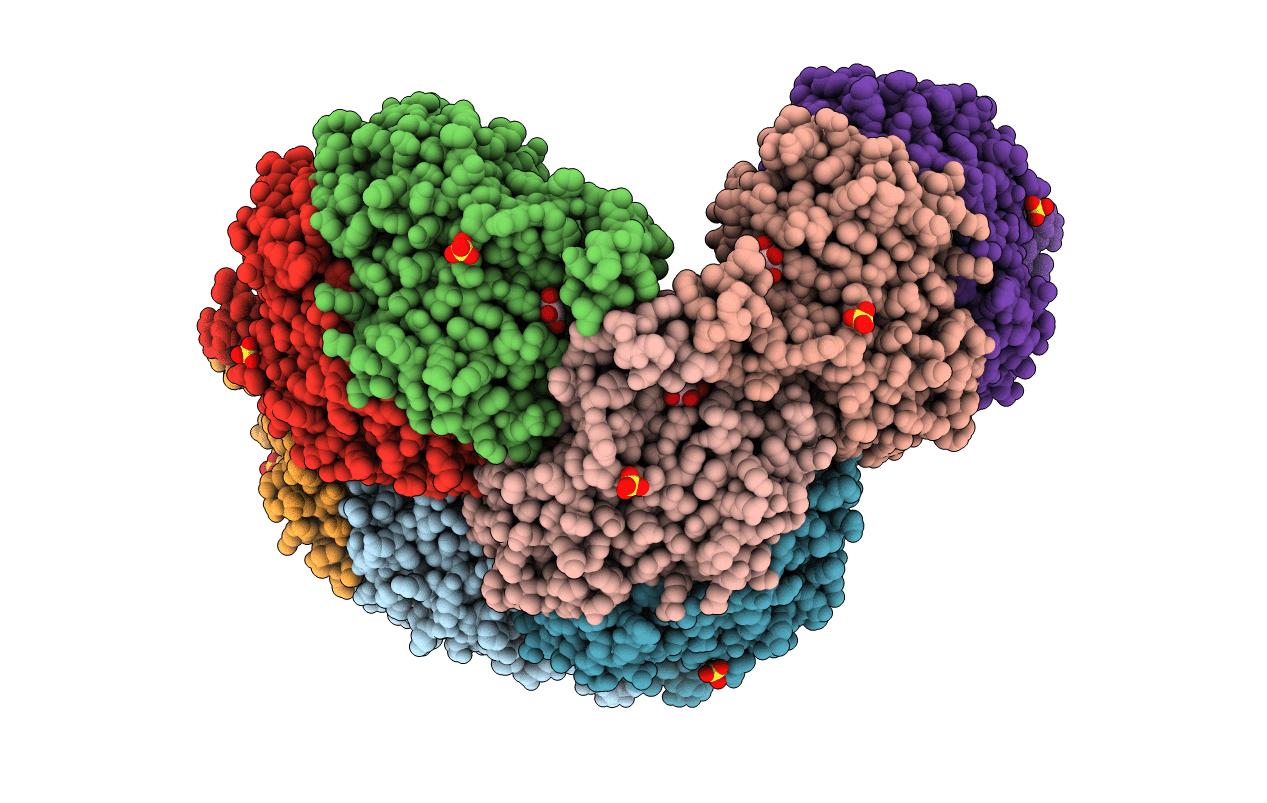 |
Crystal Structure Of Sulfur Oxygenase Reductase From Sulfurisphaera Tokodaii
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2020-07-15 Classification: OXIDOREDUCTASE Ligands: FE, GOL, SO4 |
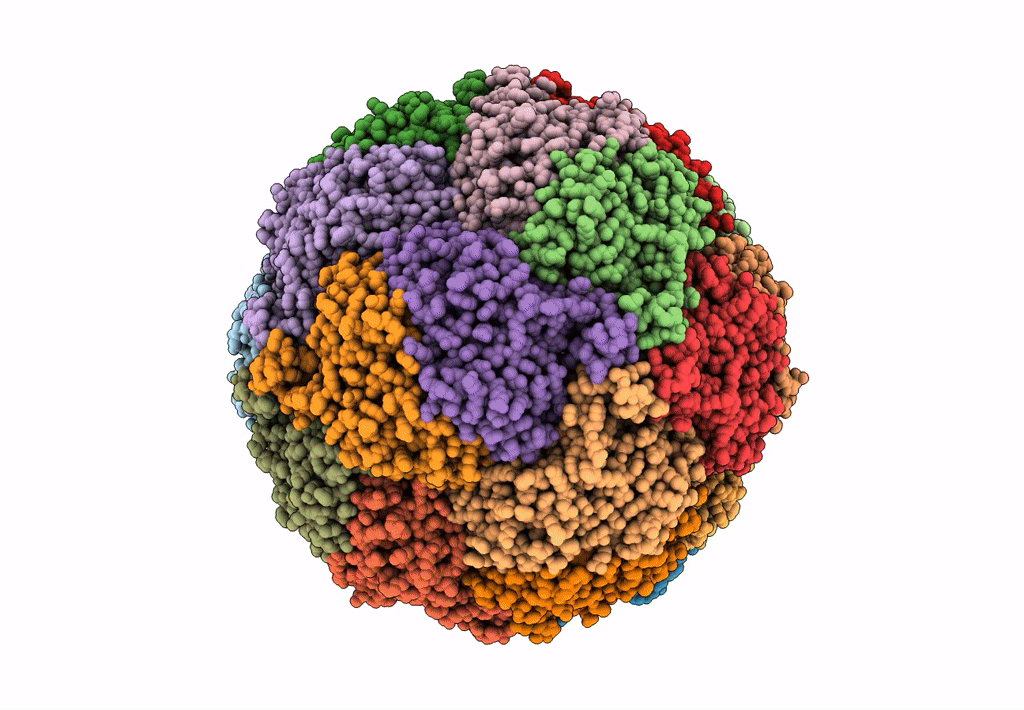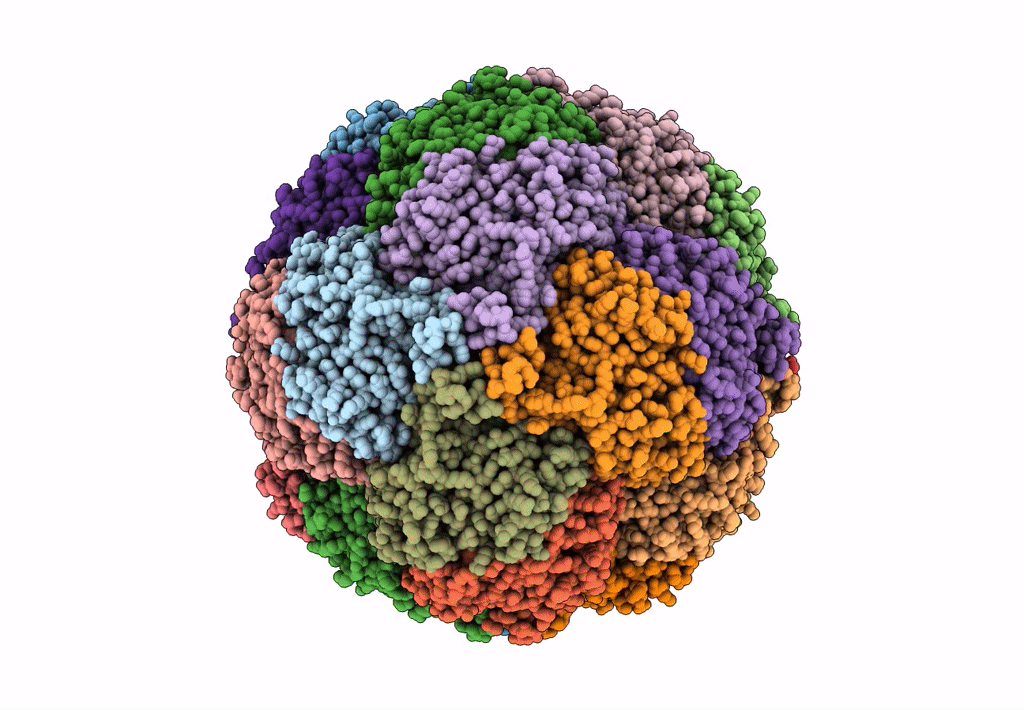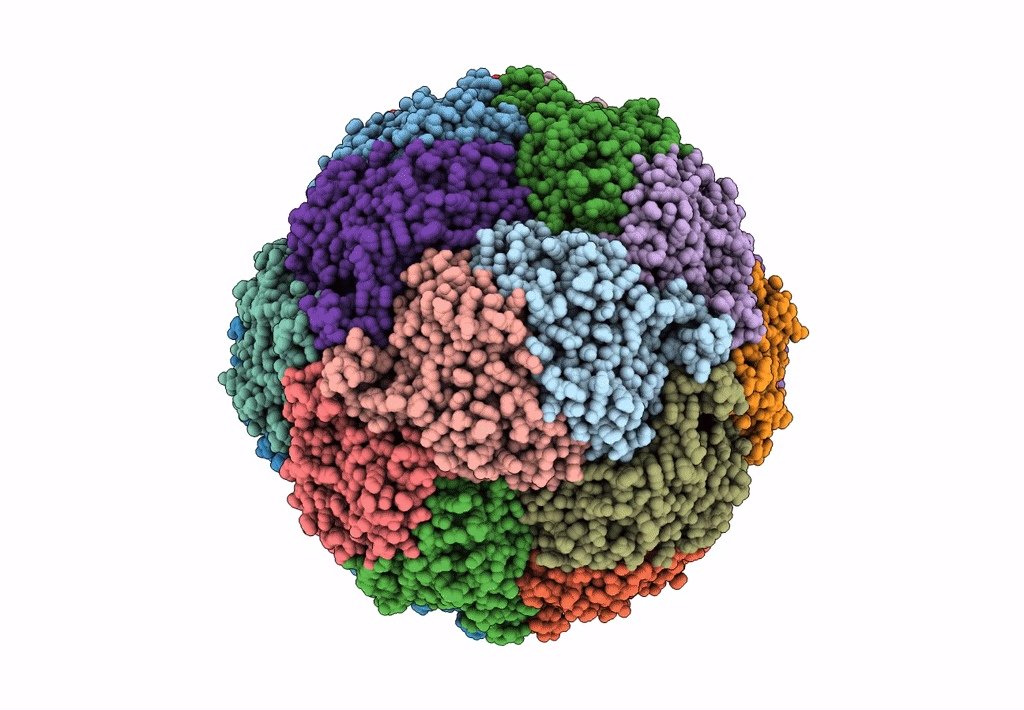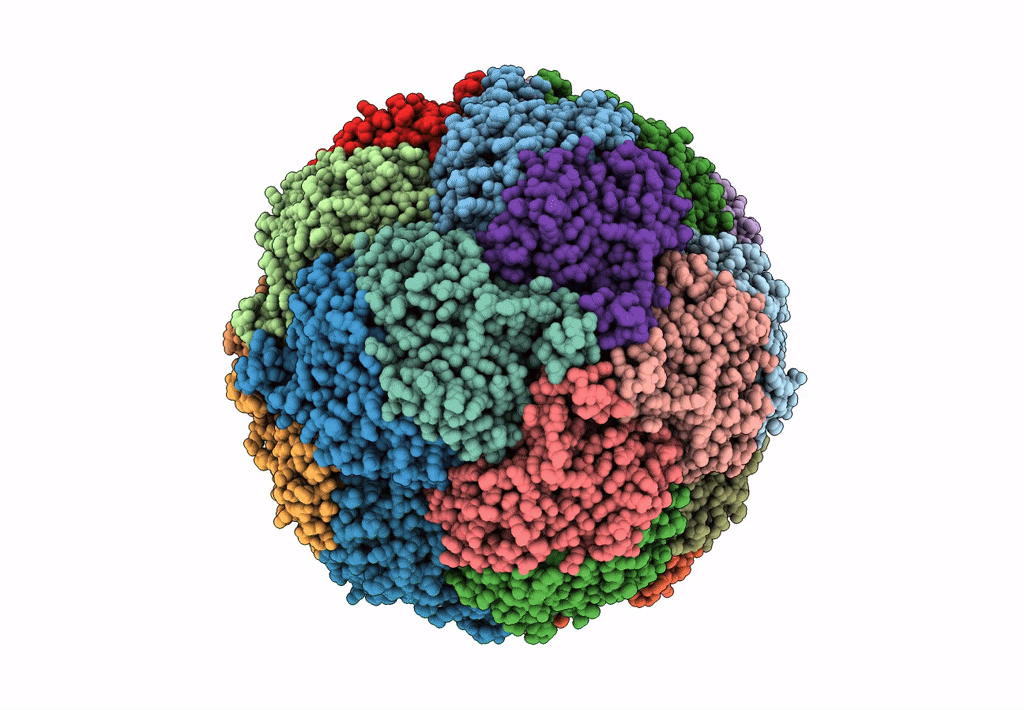 |
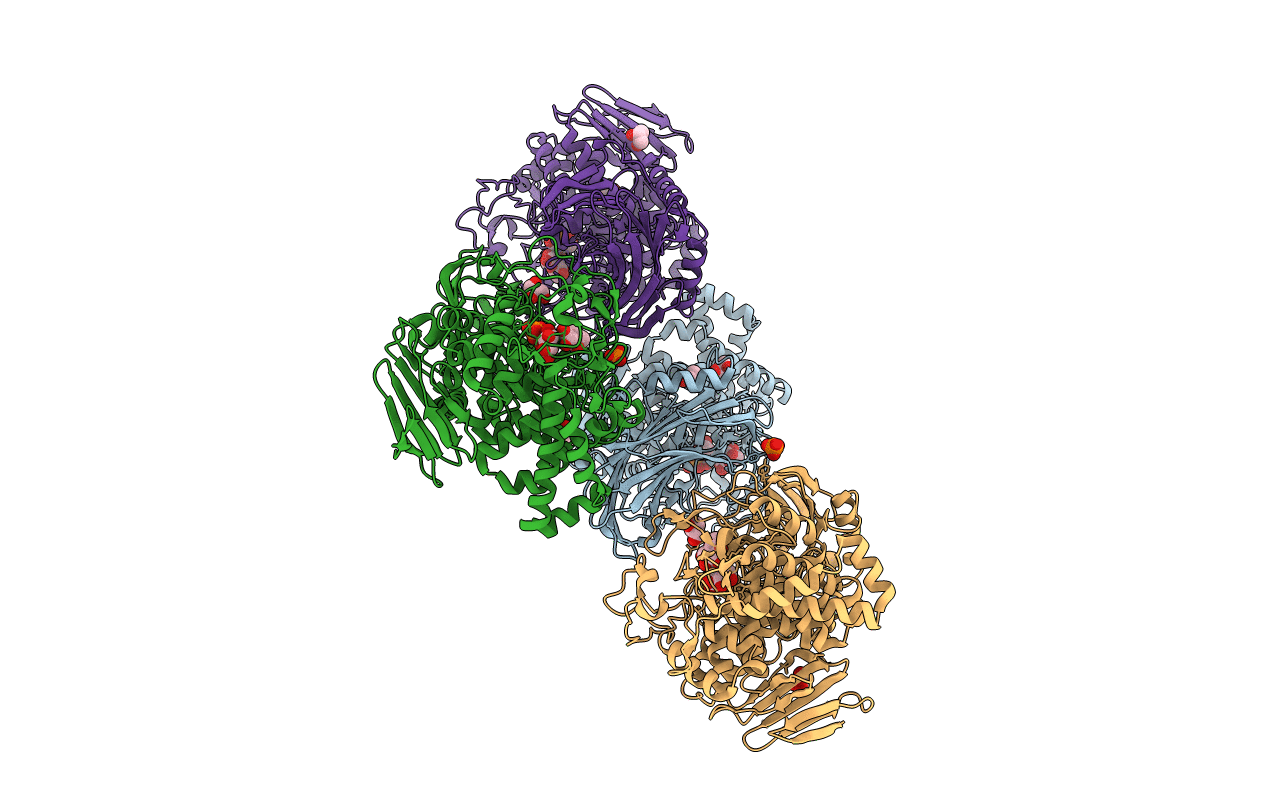
Cryo-Em Structure Of Sulfur Oxygenase Reductase From Sulfurisphaera Tokodaii
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: OXIDOREDUCTASE Ligands: FE |
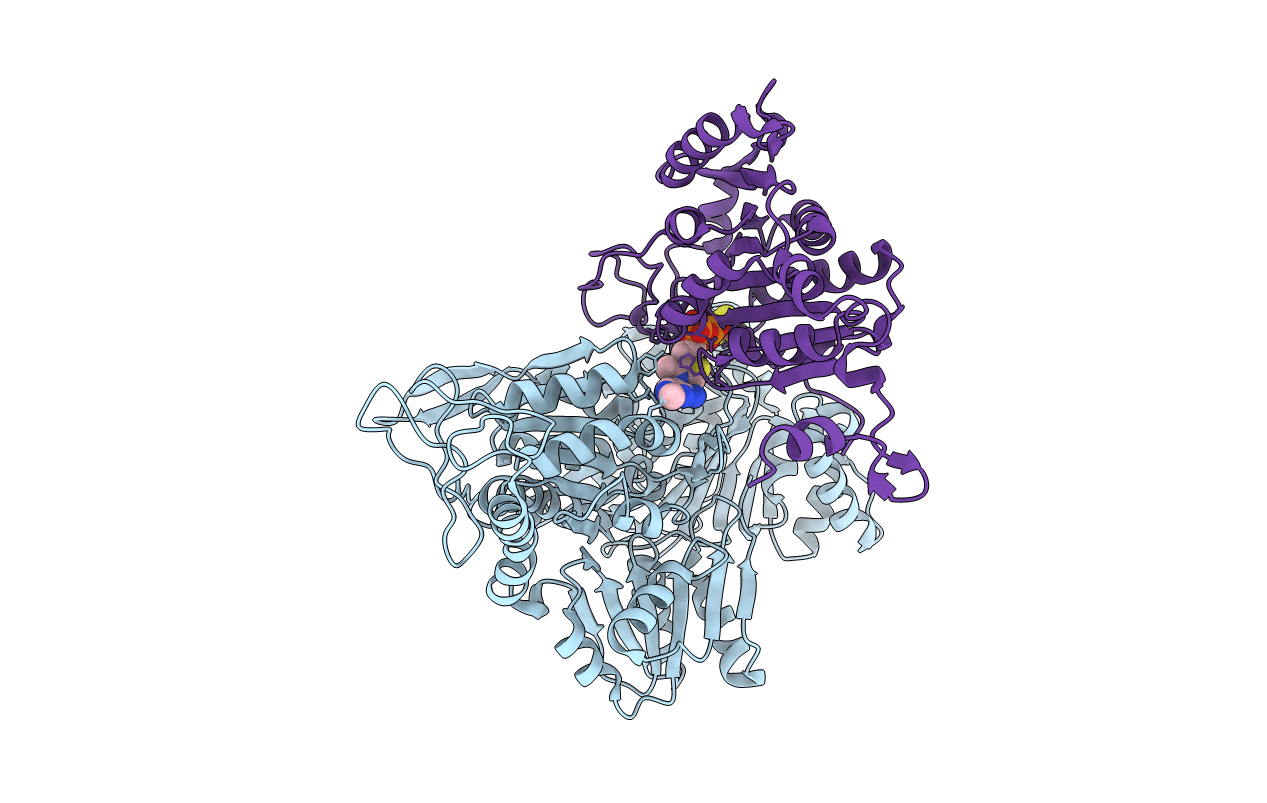 |
2-Oxoacid:Ferredoxin Oxidoreductase 2 From Sulfolobus Tokodai - Ligand Free Form
Organism: Sulfolobus tokodaii str. 7
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE Ligands: SF4, TPP, MG |
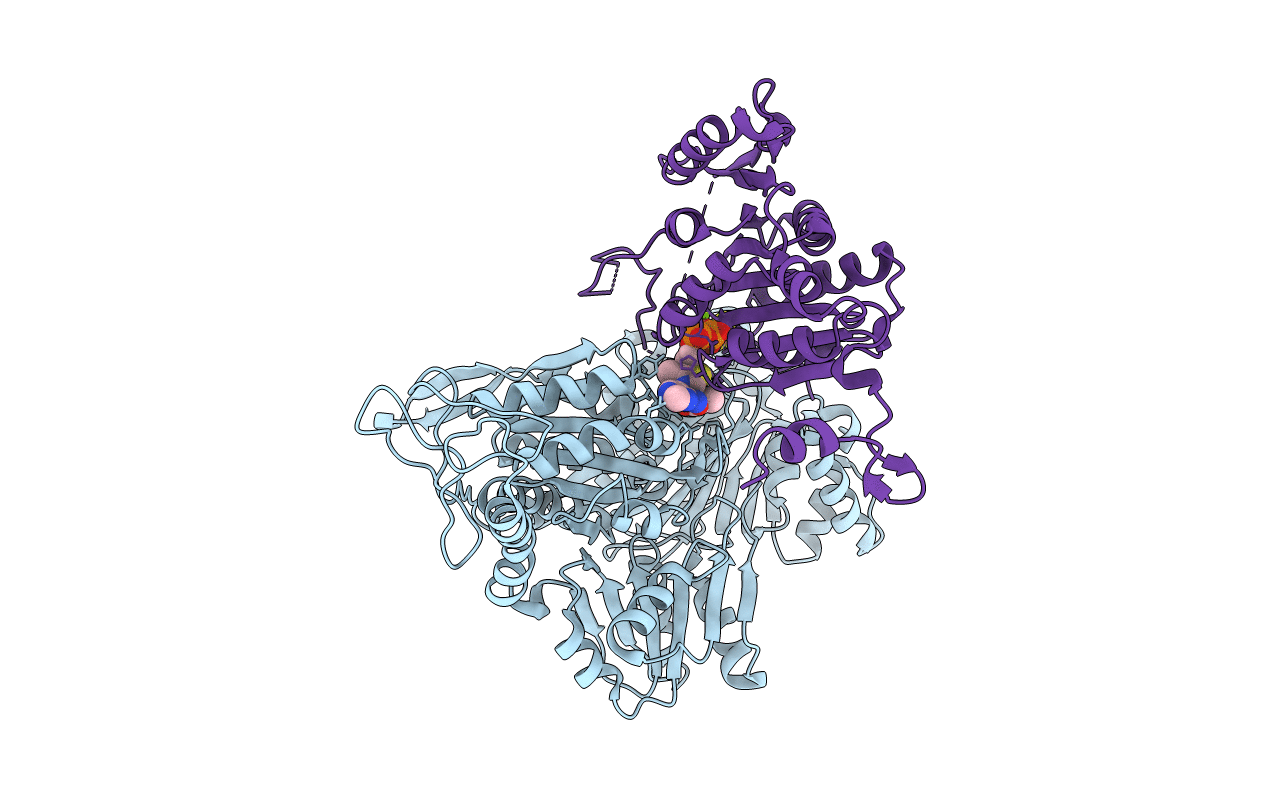 |
2-Oxoacid:Ferredoxin Oxidoreductase 2 From Sulfolobus Tokodai - Pyruvate Complex
Organism: Sulfolobus tokodaii str. 7
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE Ligands: PYR, SF4, TPP, MG |
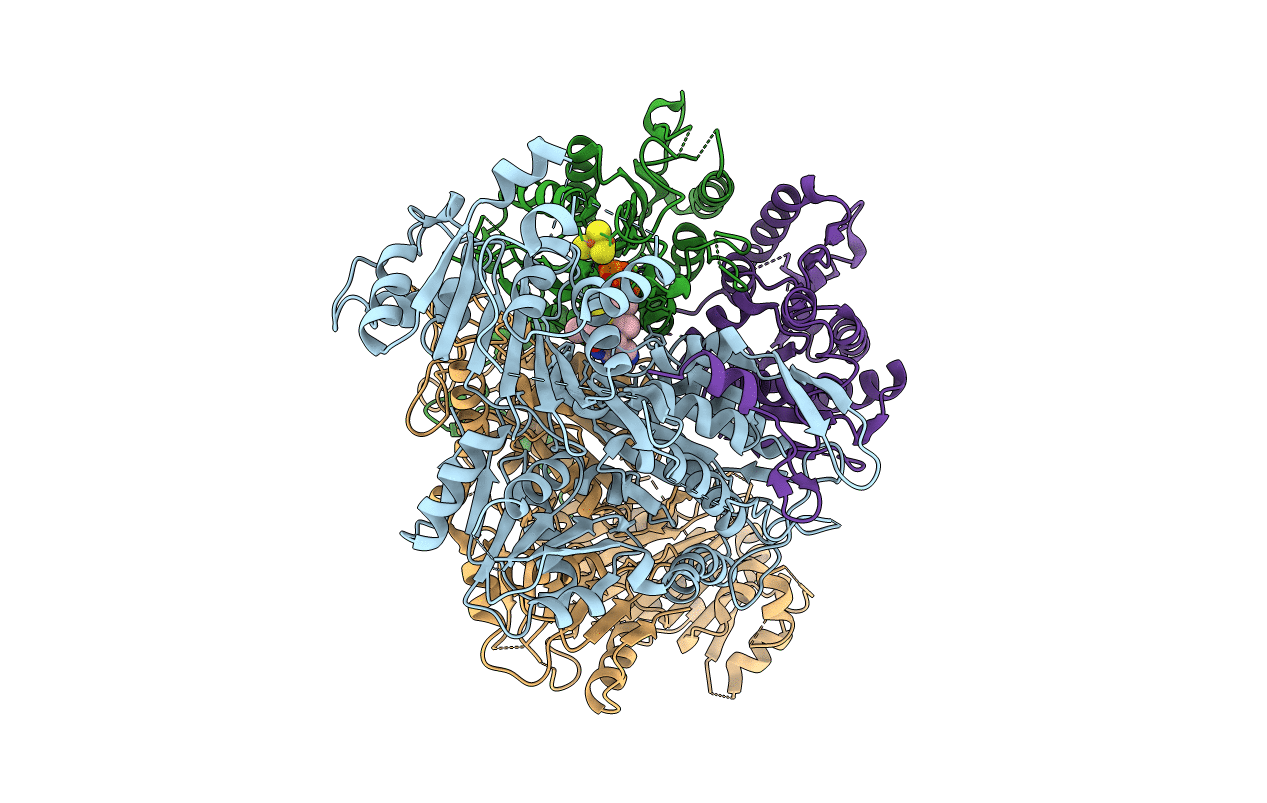 |
Organism: Sulfolobus tokodaii str. 7
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE Ligands: SF4, TDN, MG |
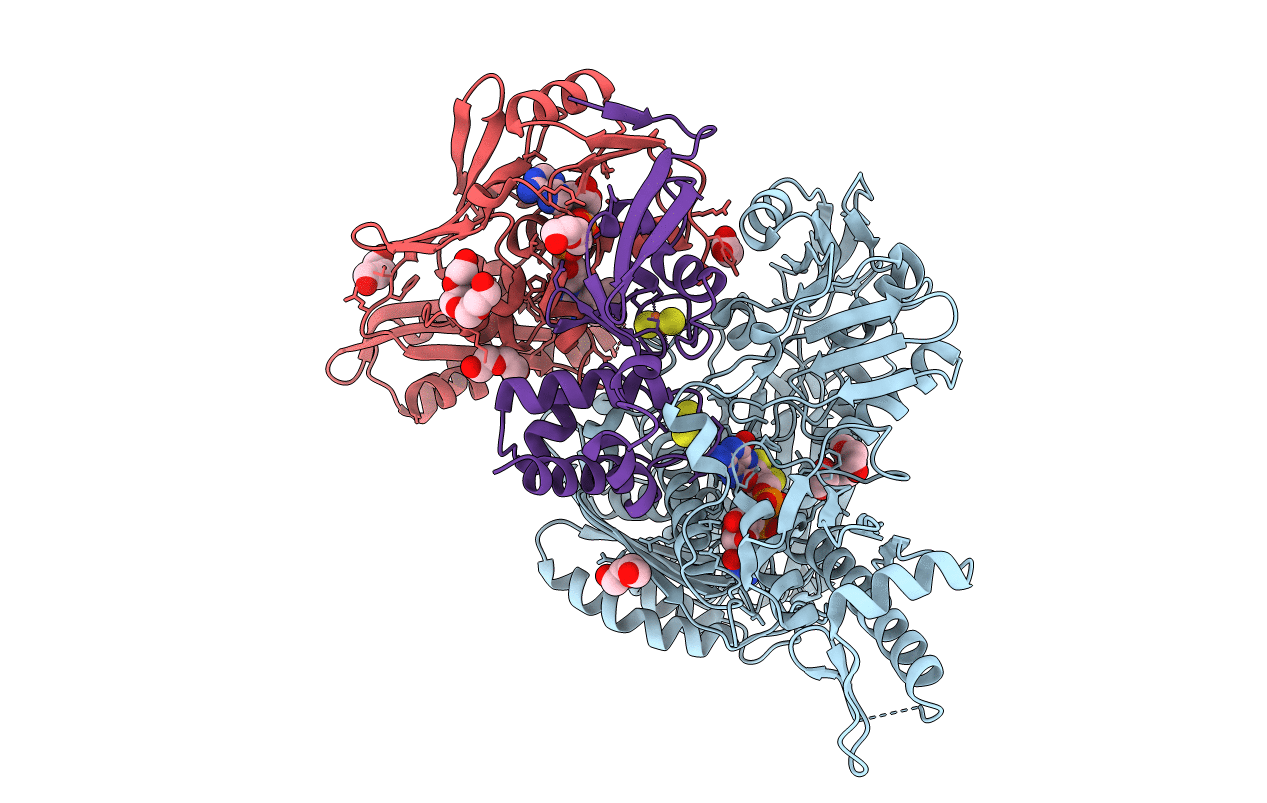 |
Organism: Sulfolobus tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-02-03 Classification: OXIDOREDUCTASE Ligands: MCN, MO, 1PE, PEG, FAD, PG4, ACY, FES |
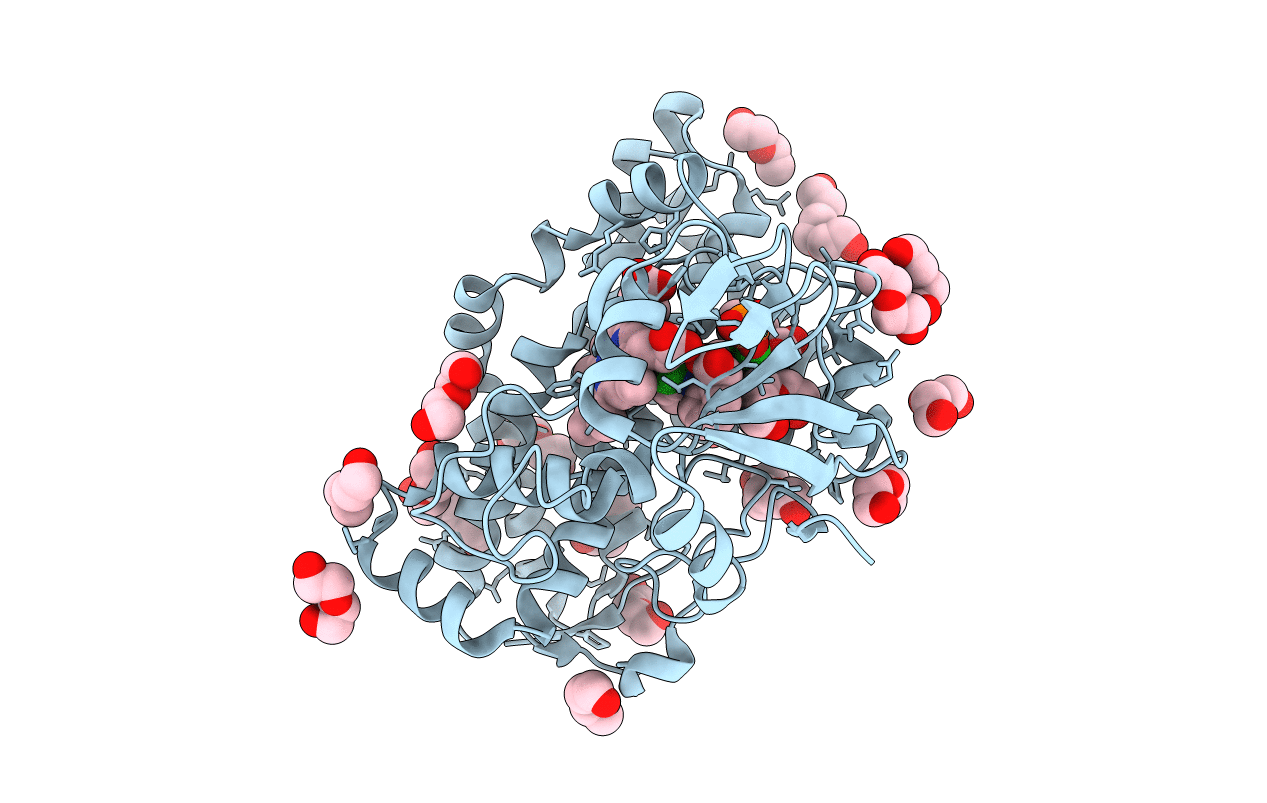 |
The Crystal Structure Of Cytochrome P450 105D7 From Streptomyces Avermitilis In Complex With Diclofenac
Organism: Streptomyces avermitilis ma-4680 = nbrc 14893
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-11-05 Classification: OXIDOREDUCTASE Ligands: HEM, DIF, PGE, PEG, PO4 |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-08-06 Classification: HYDROLASE Ligands: NAG, CA |
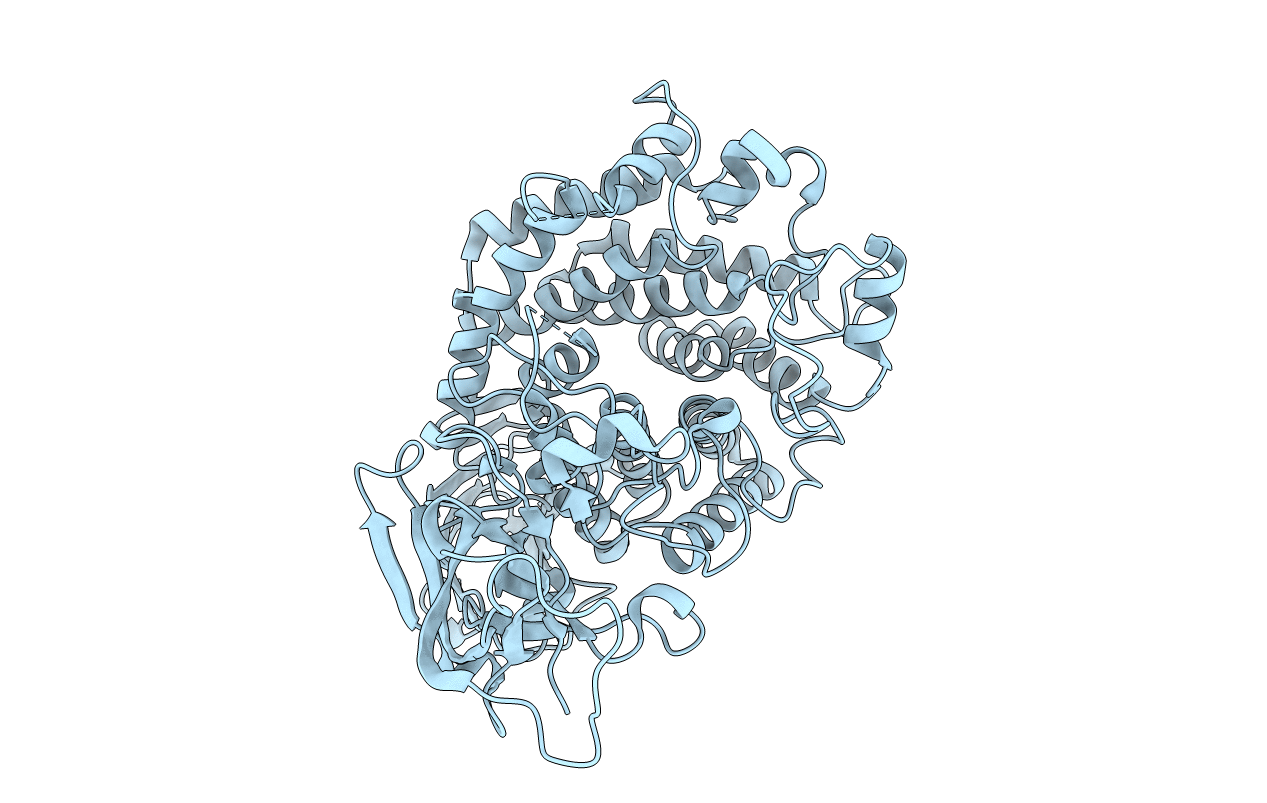 |
Crystal Structure Of Gh127 Beta-L-Arabinofuranosidase Hypba1 From Bifidobacterium Longum Ligand Free Form
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-04-30 Classification: HYDROLASE |
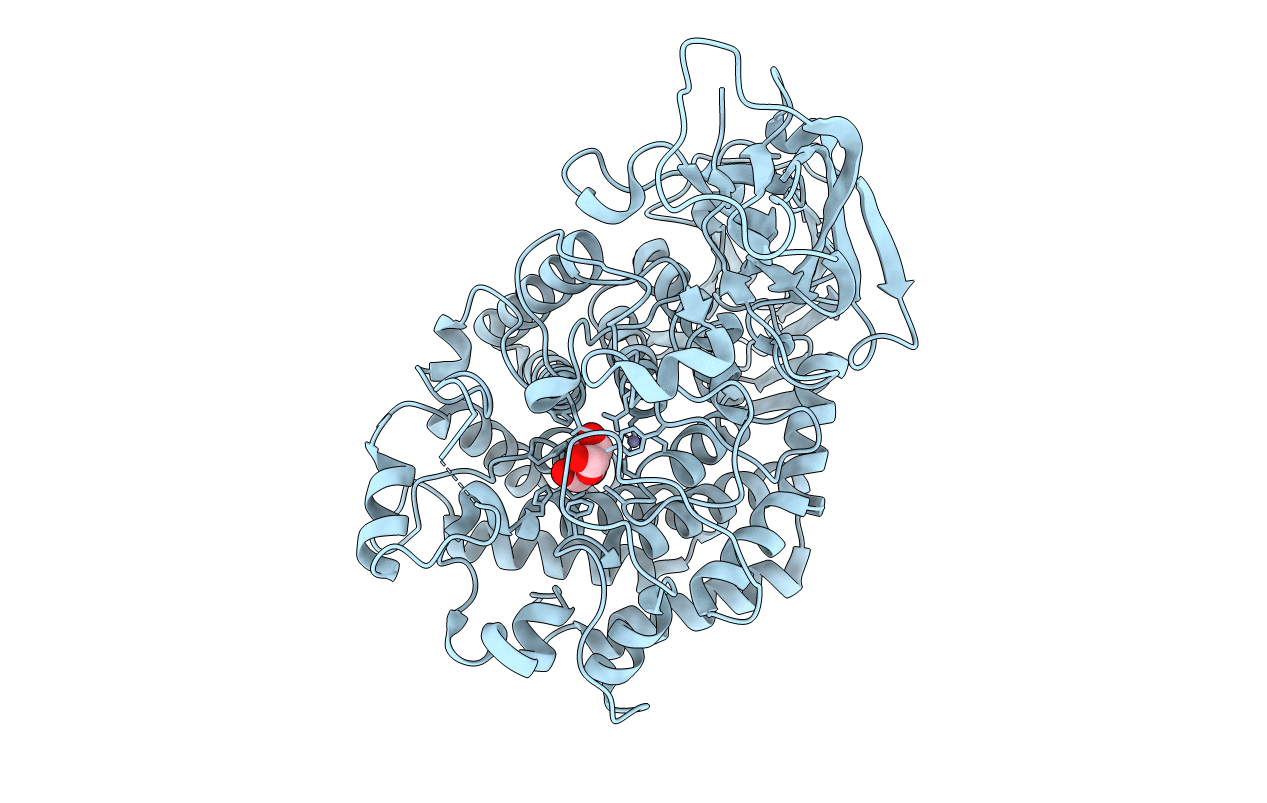 |
Crystal Structure Of Gh127 Beta-L-Arabinofuranosidase Hypba1 From Bifidobacterium Longum Arabinose Complex Form
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-04-30 Classification: HYDROLASE Ligands: FUB, ZN |
 |
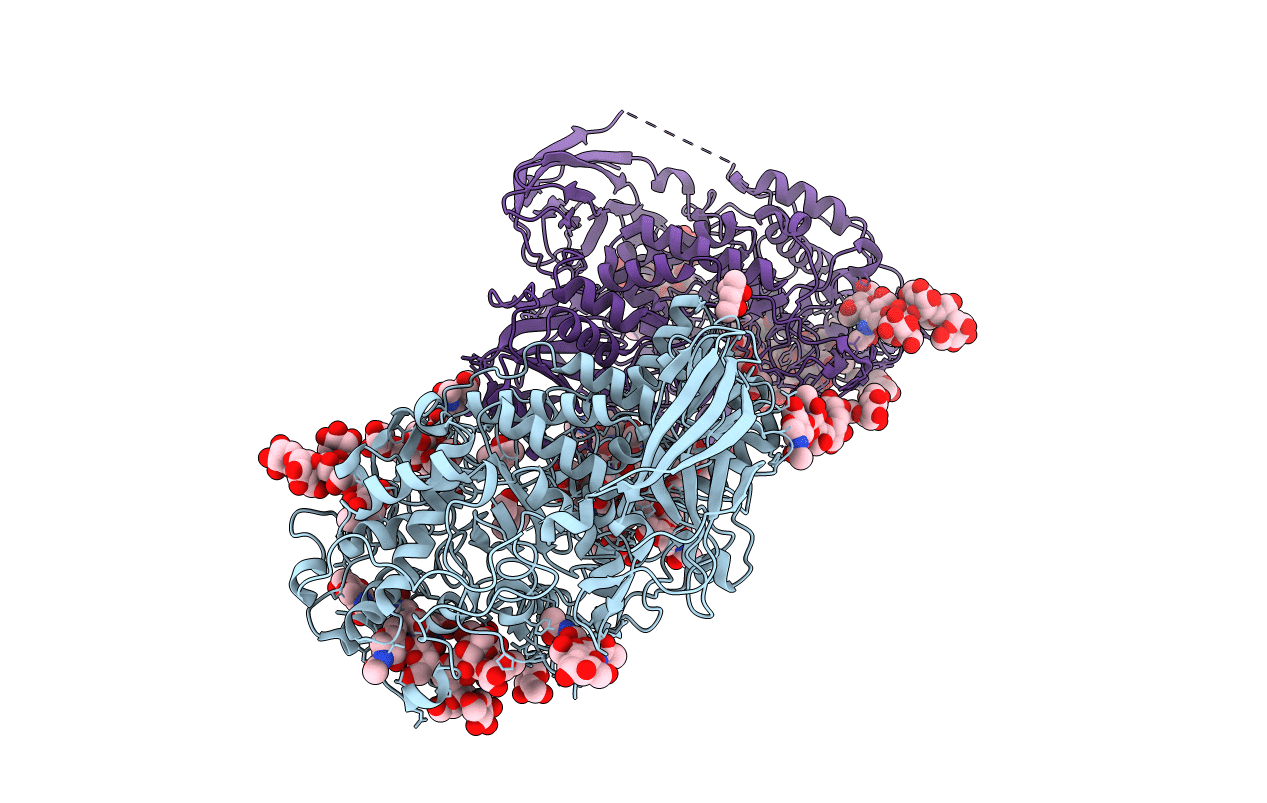
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-02-05 Classification: TRANSFERASE Ligands: SO4 |
 |
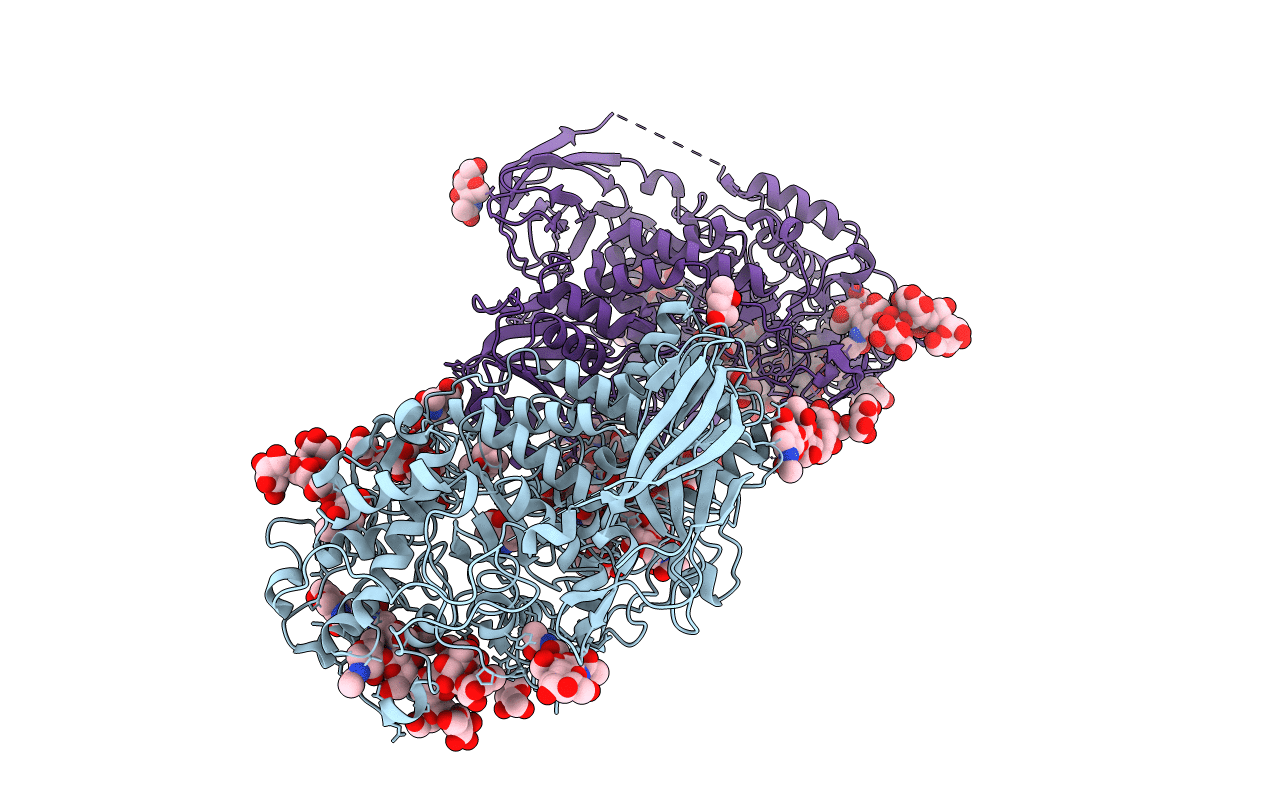
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-02-05 Classification: TRANSFERASE Ligands: BGC, GOL, PO4 |
 |
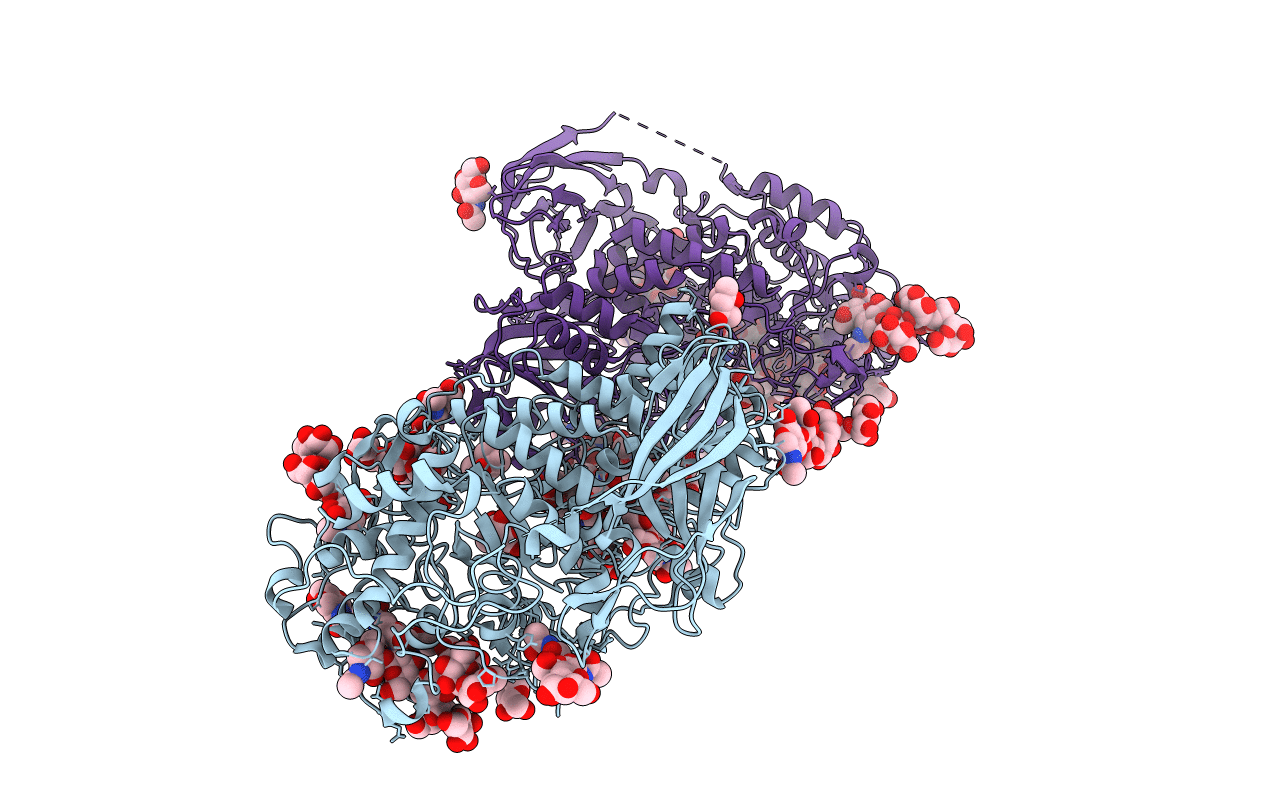
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-04-10 Classification: HYDROLASE Ligands: NAG, MPD, MRD, NA, ACY |
 |
Crystal Structure Of Beta-Glucosidase 1 From Aspergillus Aculeatus In Complex With Isofagomine
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-04-10 Classification: HYDROLASE Ligands: NAG, MRD, MPD, NA, IFM |
 |
Crystal Structure Of Beta-Glucosidase 1 From Aspergillus Aculeatus In Complex With 1-Deoxynojirimycin
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-04-10 Classification: HYDROLASE Ligands: NAG, MRD, NOJ, MPD |
 |
Crystal Structure Of Beta-Glucosidase 1 From Aspergillus Aculeatus In Complex With Calystegine B(2)
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-04-10 Classification: HYDROLASE Ligands: NAG, MRD, NA, CGB |
 |
Crystal Structure Of Beta-Glucosidase 1 From Aspergillus Aculeatus In Complex With Castanospermine
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-04-10 Classification: HYDROLASE Ligands: NAG, MRD, MPD, CTS |
 |
Crystal Structure Of Beta-Glucosidase 1 From Aspergillus Aculeatus In Complex With D-Glucose
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-04-10 Classification: HYDROLASE Ligands: NAG, MRD, BGC |
 |
Crystal Structure Of Beta-Glucosidase 1 From Aspergillus Aculeatus In Complex With Thiocellobiose
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-04-10 Classification: HYDROLASE Ligands: NAG, MRD |
 |
Organism: Bifidobacterium bifidum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-03-20 Classification: HYDROLASE Ligands: SO4 |

