Search Count: 207
 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 1
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NFU, NA, SO4, GOL, FE, SF4, FAD, CL, EPE |
 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 2
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NFU, GOL, FE, SF4, FAD |
 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 3
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: GOL, MES, NFU, FE, SO4, EDO, FAD, SF4, PE4, FES |
 |
Cubic State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: SO4, NFU, FAD, GOL, SF4 |
 |
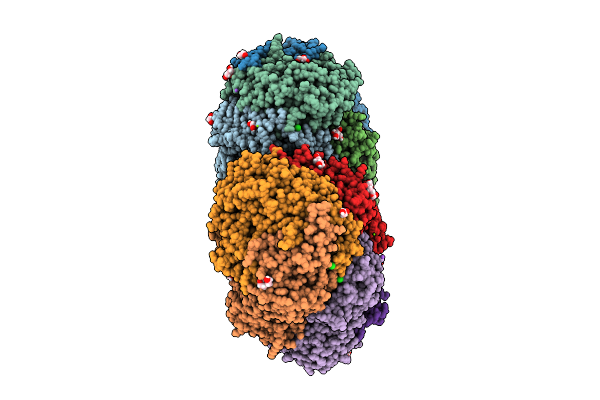
Methyl-Coenzyme M Reductase Of Anme-2D Candidatus Methanoperedens Vercelli Strain 1 From A Bioreactor Enrichment Culture
Organism: Candidatus methanoperedens sp.
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: K, F43, EDO, TP7, COM, SHT |
 |
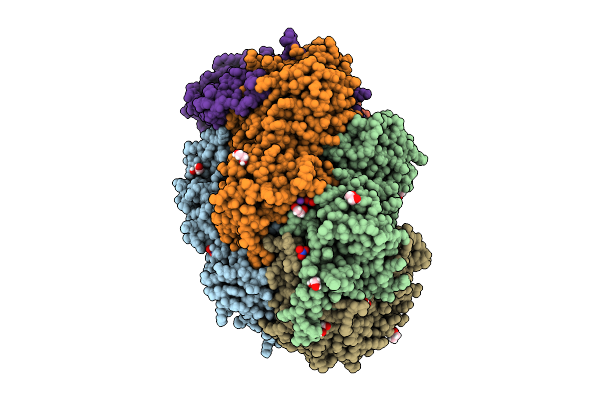
Krypton-Pressurized Methyl-Coenzyme M Reductase Of An Anme-2C Isolated From A Microbial Enrichment
Organism: Candidatus methanogasteraceae archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: KR, MG, PGE, EDO, CL, TP7, F43, PEG, GOL, COM, NA, K |
 |
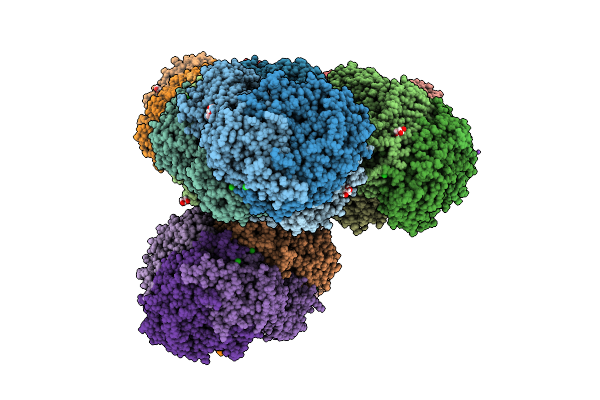
Methyl-Coenzyme M Reductase Of Anme-2D Candidatus Methanoperedens Sp. Blz2 From A Bioreactor Enrichment Culture
Organism: Candidatus methanoperedens sp. blz2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE |
 |
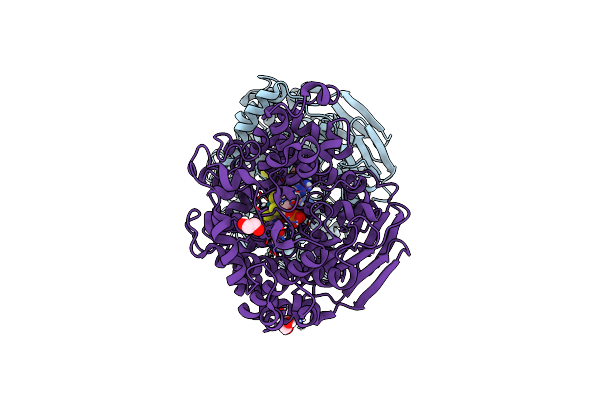
Organism: Candidatus methanogasteraceae archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: K, NA, CL, F43, COM, TP7, GOL, MPD, EDO |
 |
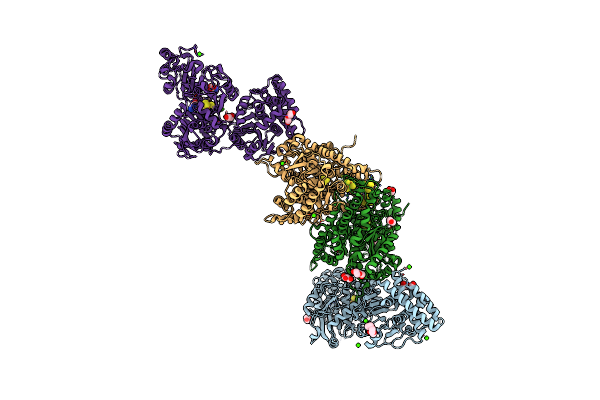
Crystal Structure Of The Tungsten-Dependent Aldehyde:Ferredoxin Oxidoreductase From Clostridium Autoethanogenum.
Organism: Clostridium autoethanogenum dsm 10061
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: SF4, A1IJH, CL, MG, EDO |
 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Acetyl-Coenyzme A From Clostridium Autoethanogenum
Organism: Clostridium autoethanogenum dsm 10061
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: SF4, NI, EDO, PE4, GOL, PEG, CA, CL, XCC, ACO, TRS |
 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3A)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, B12, NI, RQM |
 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3B)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, RQM, B12 |
 |
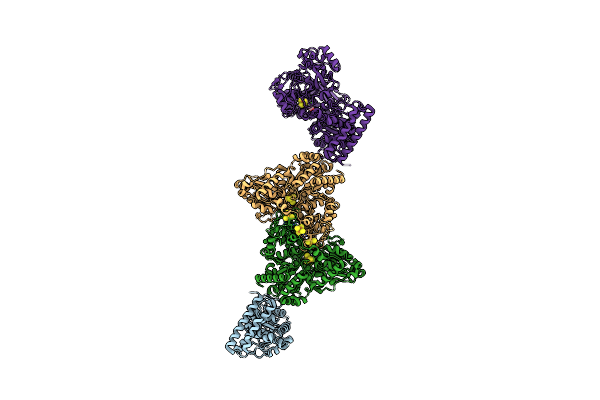
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3Cb)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, B12, RQM |
 |
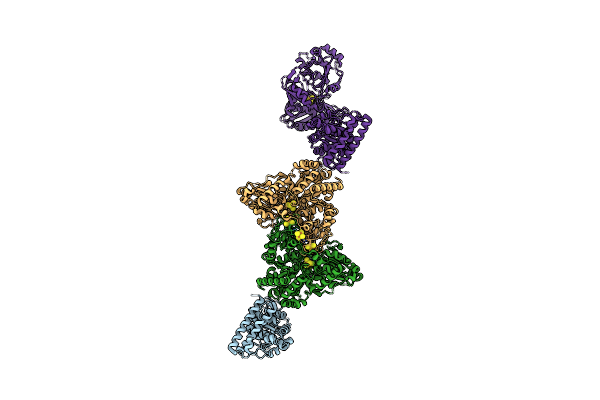
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) From Clostridium Autoethanogenum (Composite Structure, Closed And Co-Bound State)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, RQM, NI, CMO |
 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) From Clostridium Autoethanogenum (Composite Structure, Semi-Extended State)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, RQM |
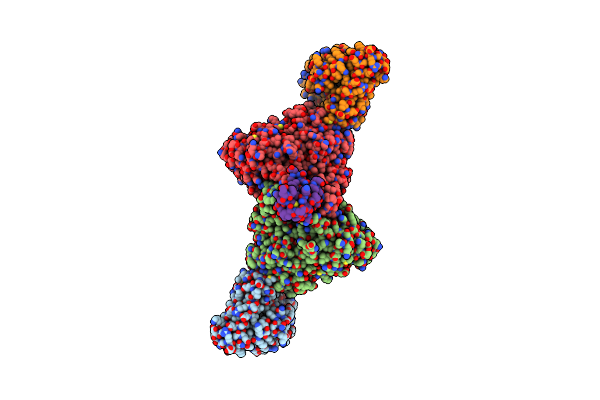 |
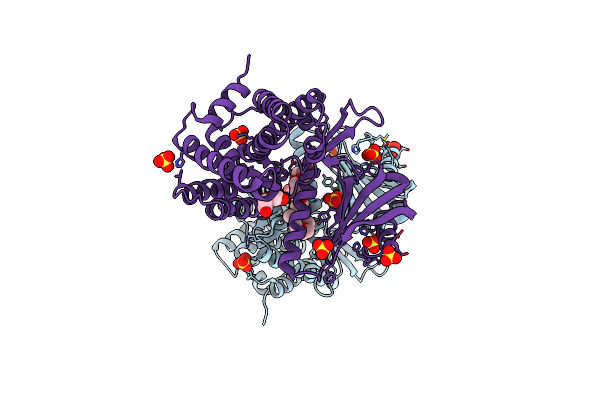
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Ferredoxin (Clostridium Autoethanogenum)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, RQM |
 |
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2024-11-06 Classification: TRANSFERASE Ligands: PEG, SO4, CL, RWB |
 |
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2024-10-23 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH, XP6, CL |
 |
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2024-10-23 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH, XP6, CL |
 |
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2024-10-23 Classification: BIOSYNTHETIC PROTEIN Ligands: SFG, XPN, CL |

