Search Count: 52
 |
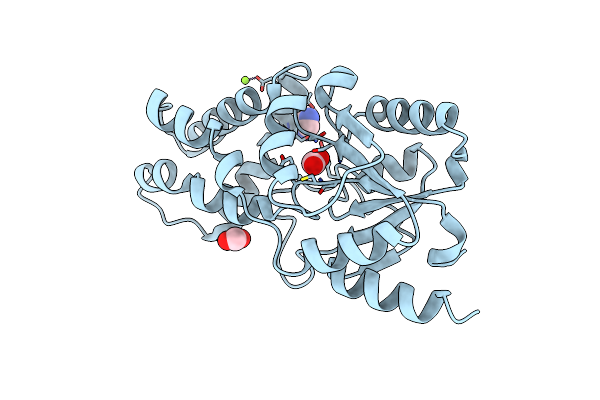
Crystal Structure Of The Enzyme-Product Complex Of L-Azetidine-2-Carboxylate Hydrolase
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:0.93 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: 42B, IMD, FMT, EDO, PEG, MG |
 |
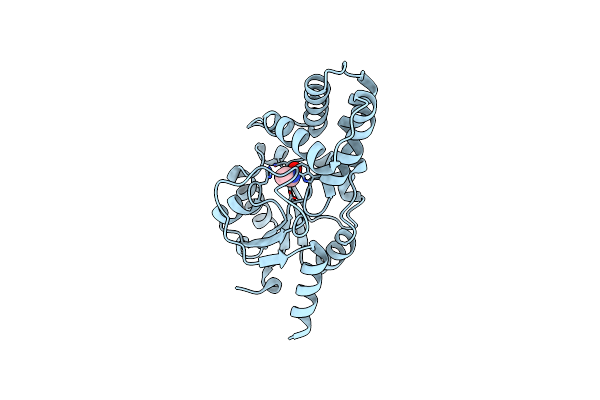
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: FMT, IMD, MG |
 |
Crystal Structure Of L-Azetidine-2-Carboxylate Hydrolase Soaked In (S)-Azetidine-2-Carboxylic Acid
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: 02A |
 |
Crystal Structure Of Metformin Hydrolase (Mfmab) From Pseudomonas Mendocina Sp. Met-2 With Ni2+2 Bound
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NI |
 |
Crystal Structure Of Metformin Hydrolase (Mfmab) From Pseudomonas Mendocina Sp. Met-2 Mutant (Mfma/D188N)
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Metformin Hydrolase (Mfmab) From Pseudomonas Mendocina Sp. Met-2 Apo Form
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NI |
 |
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: BME |
 |
Triuret Hydrolase (Trta) From Herbaspirillum Sp. Bh-1 C162S Bound With Biuret
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: EDO, C5J |
 |
Triuret Hydrolase (Trta) From Herbaspirillum Sp. Bh-1 C162S Bound With Triuret
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: TIU, EDO |
 |
Organism: Rhodococcus sp. mel
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: C5J |
 |
Organism: Rhodococcus sp. mel
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-18 Classification: HYDROLASE |
 |
Organism: Moorella thermoacetica atcc 39073
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: MLI, CA, PDO, CL |
 |
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: MLI, CA, CL, PDO |
 |
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: MLI, CA, PDO |
 |
Crystal Structures Of Cyanuric Acid Hydrolase From Moorella Thermoacetica Complexed With Cyanuric Acid
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: MLI, CA, PDO, PEG, WDL, CL |
 |
Crystal Structures Of Cyanuric Acid Hydrolase From Moorella Thermoacetica Complexed With Barbituric Acid
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: MLI, CA, NA, PDO, CL, BR8, PEG |
 |
Crystal Structures Of Cyanuric Acid Hydrolase From Moorella Thermoacetica Complexed With Barbituric Acid
Organism: Moorella thermoacetica atcc 39073
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: BR8, MLI, CA |
 |
Crystal Structures Of Cyanuric Acid Hydrolase From Moorella Thermoacetica Complexed With 1,3-Acetone Dicarboxylic Acid
Organism: Moorella thermoacetica (strain atcc 39073 / jcm 9320)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: MLI, CA, ACT, 6JN, PDO |
 |
Organism: Xanthomonas campestris pv. campestris (strain atcc 33913 / dsm 3586 / ncppb 528 / lmg 568 / p 25)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: GOL |

