Search Count: 41
 |
Crystal Structure Of M- And C-Domains Of The Shaft Pilin Lrpa From Ligilactobacillus Ruminis - Iodide Derivative
Organism: Ligilactobacillus ruminis atcc 25644
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2024-07-10 Classification: CELL ADHESION Ligands: IOD, NA, CL, EDO, GOL |
 |
Crystal Structure Of M- And C-Domains Of The Shaft Pilin Lrpa From Ligilactobacillus Ruminis - Triclinic Form
Organism: Ligilactobacillus ruminis atcc 25644
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-07-10 Classification: CELL ADHESION |
 |
Crystal Structure Of M- And C-Domains Of The Shaft Pilin Lrpa From Ligilactobacillus Ruminis - Orthorhombic Form
Organism: Ligilactobacillus ruminis atcc 25644
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-07-10 Classification: CELL ADHESION Ligands: IOD, NA, GOL, EDO |
 |
Organism: Ligilactobacillus ruminis atcc 25644
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-07-10 Classification: CELL ADHESION Ligands: IOD |
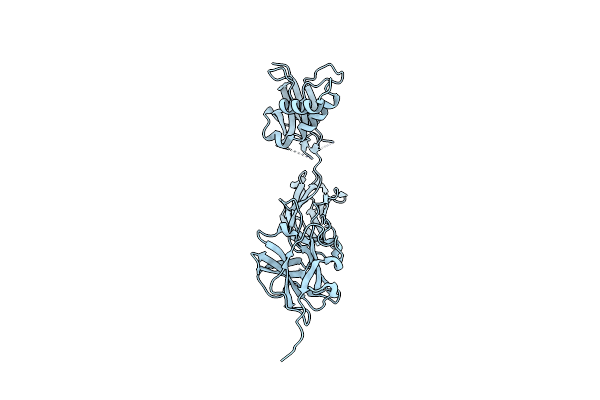 |
Crystal Structure Of The Shaft Pilin Lrpa From Ligilactobacillus Ruminis - Orthorhombic Form
Organism: Ligilactobacillus ruminis atcc 25644
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-10 Classification: CELL ADHESION |
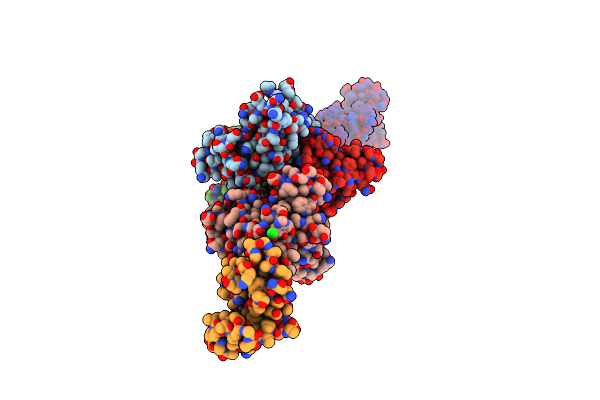 |
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2020-12-30 Classification: CELL ADHESION Ligands: MG, CL |
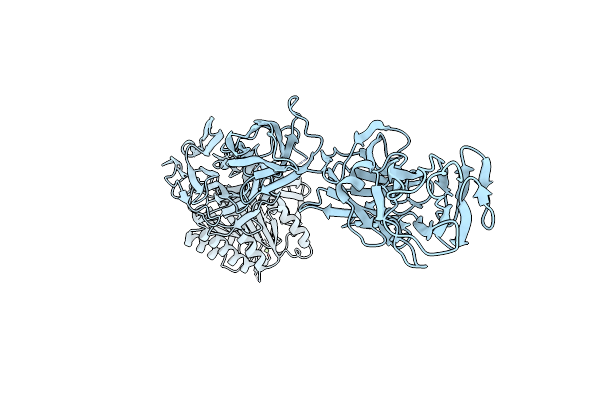 |
Crystal Structure Of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - Open Conformation
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-07-29 Classification: CELL ADHESION Ligands: MG |
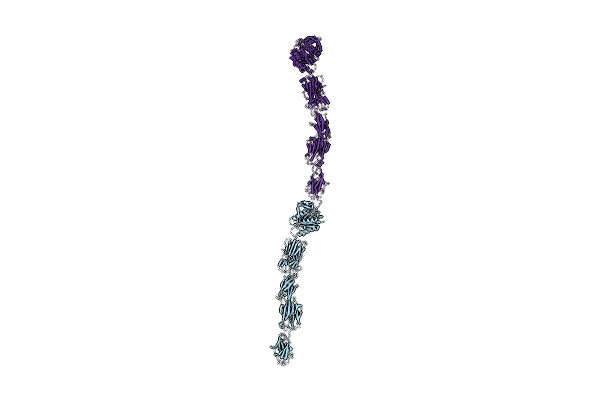 |
Crystal Structure Of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - P21212 Form
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-07-29 Classification: CELL ADHESION Ligands: MG, CL |
 |
Crystal Structure Of C-Terminal Fragment Of Pilus Adhesin Spac From Lactobacillus Rhamnosus Gg
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2020-07-29 Classification: CELL ADHESION |
 |
Crystal Structure Of C-Terminal Fragment Of Pilus Adhesin Spac From Lactobacillus Rhamnosus Gg-Iodide Soaked
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2020-07-29 Classification: CELL ADHESION Ligands: IOD |
 |
Crystal Structure Of Spae Basal Pilin From Lactobacillus Rhamnosus Gg - Selenium Derivative
Organism: Lactobacillus rhamnosus (strain atcc 53103 / gg)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2019-06-26 Classification: CELL ADHESION |
 |
Crystal Structure Of Spae Basal Pilin From Lactobacillus Rhamnosus Gg - Orthorhombic Form
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2019-06-26 Classification: CELL ADHESION |
 |
Crystal Structure Of Spae Basal Pilin From Lactobacillus Rhamnosus Gg - Trigonal Form
Organism: Lactobacillus rhamnosus (strain atcc 53103 / gg)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-06-26 Classification: CELL ADHESION |
 |
Organism: lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2018-06-20 Classification: CELL ADHESION |
 |
Crystal Structure Of C-Terminal Fragment Of Spad From Lactobacillus Rhamnosus Gg Generated By Limited Proteolysis
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2018-06-20 Classification: CELL ADHESION Ligands: CL |
 |
Crystal Structure Of Shaft Pilin Spad From Lactobacillus Rhamnosus Gg In Bent Conformation
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2018-06-20 Classification: CELL ADHESION |
 |
Crystal Structure Of Shaft Pilin Spad From Lactobacillus Rhamnosus Gg - D242A Mutant
Organism: lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2018-06-20 Classification: CELL ADHESION |
 |
Crystal Structure Of Shaft Pilin Spad From Lactobacillus Rhamnosus Gg - K365A Mutant
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-06-20 Classification: CELL ADHESION |
 |
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-07-20 Classification: CELL ADHESION Ligands: ACT |
 |
Crystal Structure Of C-Terminal Domain Of Shaft Pilin Spaa From Lactobacillus Rhamnosus Gg, - I422 Space Group
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-07-20 Classification: CELL ADHESION Ligands: EDO |

