Search Count: 20
 |
Structure Of The Human 80S Ribosome At 1.9 A Resolution - The Molecular Role Of Chemical Modifications And Ions In Rna
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: RIBOSOME Ligands: MG, K, SPD, ZN |
 |
Cytochrome Bcc-Aa3 Supercomplex (Respiratory Supercomplex Iii2/Iv2) From Corynebacterium Glutamicum (Stigmatellin And Azide Bound)
|
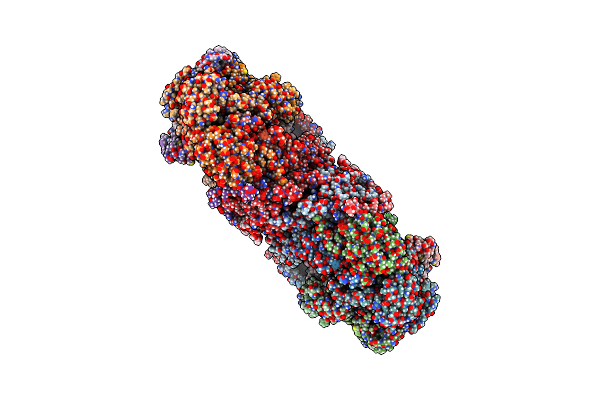 |
Cytochrome Bcc-Aa3 Supercomplex (Respiratory Supercomplex Iii2/Iv2) From Corynebacterium Glutamicum (As Isolated)
|
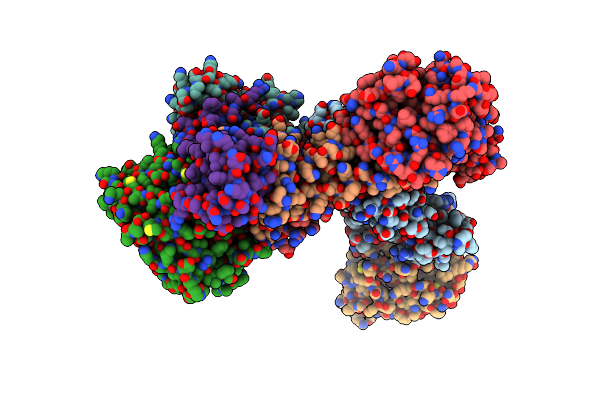 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: STRUCTURAL PROTEIN Ligands: ATP |
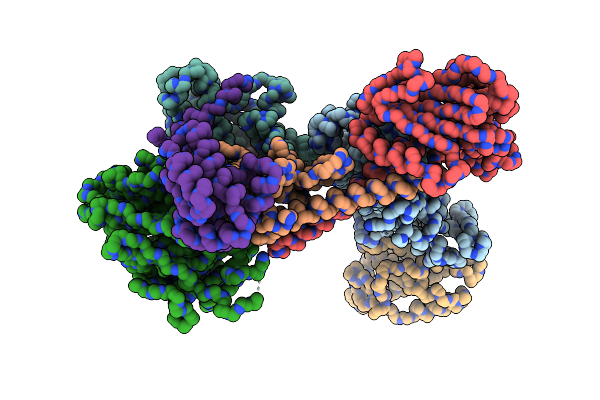 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: STRUCTURAL PROTEIN Ligands: ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: RIBOSOME Ligands: MG, 3HE, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: RIBOSOME Ligands: MG, 3HE, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: RIBOSOME Ligands: MG, ZN |
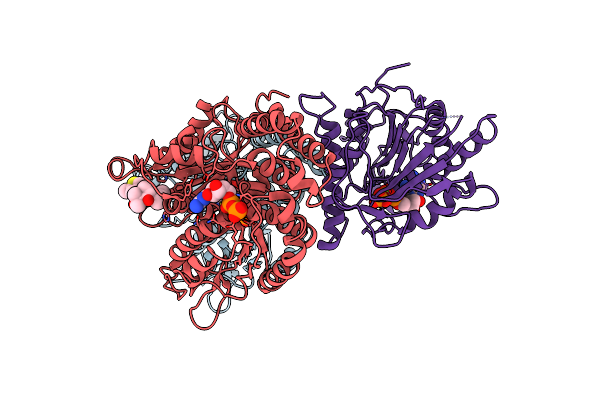 |
Organism: Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2019-08-21 Classification: MOTOR PROTEIN Ligands: MG, ANP, GDP, EPB, GTP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2019-02-13 Classification: GENE REGULATION Ligands: ZN, GOL |
 |
Ustilago Maydis Kinesin-5 Motor Domain With N-Terminal Extension In The Amppnp State Bound To Microtubules
Organism: Ustilago maydis, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2018-08-08 Classification: MOTOR PROTEIN Ligands: MG, ANP, GTP, TA1, GDP |
 |
Ustilago Maydis Kinesin-5 Motor Domain In The Amppnp State Bound To Microtubules
Organism: Ustilago maydis (strain 521 / fgsc 9021), Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2018-06-27 Classification: MOTOR PROTEIN Ligands: MG, ANP, GTP, TA1, GDP |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2017-12-20 Classification: STRUCTURAL PROTEIN Ligands: GDP, GTP |
 |
Pseudo-Atomic Model Of Microtubule-Bound S.Pombe Kinesin-5 Motor Domain In The Amppnp State (Based On Cryo-Electron Microscopy Experiment): The N-Terminus Conformation Allows Formation Of A Cover Neck Bundle.
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-11-30 Classification: MOTOR PROTEIN Ligands: MG, GTP, GDP, TA1, ANP |
 |
Pseudo-Atomic Model Of Microtubule-Bound S. Pombe Kinesin-5 Motor Domain In The Amppnp State (Based On Cryo-Electron Microscopy Experiment): The N-Terminus Adopts Multiple Conformations
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-11-30 Classification: MOTOR PROTEIN Ligands: MG, GTP, GDP, TA1, ANP |
 |
Pseudo-Atomic Model Of Microtubule-Bound S.Pombe Kinesin-5 Motor Domain In The Amppnp State (Based On Cryo-Electron Microscopy Experiment): The N-Terminus Adopts Multiple Conformations.
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-11-30 Classification: MOTOR PROTEIN Ligands: MG, GTP, GDP, TA1, ANP |
 |
Pseudo-Atomic Model Of Microtubule-Bound S.Pombe Kinesin-5 Motor Domain In The Amppnp State (Based On Cryo-Electron Microscopy Experiment): The N-Terminus Adopts Multiple Conformations.
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-11-30 Classification: MOTOR PROTEIN Ligands: MG, GTP, GDP, TA1, ANP |
 |
Pseudo-Atomic Model Of Microtubule-Bound S.Pombe Kinesin-5 Motor Domain In The Amppnp State (Based On Cryo-Electron Microscopy Experiment): The N-Terminus Adopts Multiple Conformations.
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-11-30 Classification: MOTOR PROTEIN Ligands: MG, GTP, GDP, TA1, ANP |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:5.70 Å Release Date: 2015-03-25 Classification: RIBOSOME Ligands: MG |
 |
Structural Basis Of Signal Sequence Surveillance And Selection By The Srp-Sr Complex
Organism: Escherichia coli, Sulfolobus solfataricus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:12.00 Å Release Date: 2013-03-06 Classification: PROTEIN TRANSPORT Ligands: MG |

